Abstract
An antiserum raised against dehydrin from maize (Zea mays) recognised several polypeptides in extracts of pea (Pisum sativum) cotyledons. A cDNA expression library was prepared from mRNA of developing cotyledons, screened with the antiserum and positive clones were purified and characterised. The nucleotide sequence of one such clone, pPsB12, contained an open reading frame which would encode a polypeptide with regions of significant amino acid sequence similarity to dehydrins from other plant species.
The deduced amino acid sequence of the pea dehydrin encoded by B12 is 197 amino acids in length, has a high glycine content (25.9%), lacks tryptophan and is highly hydrophilic. The polypeptide has an estimated molecular mass of 20.4 kDa and pI=6.4. An in vitro synthesised product from the clone comigrates with one of the in vivo proteins recognised by the antiserum.
A comparison of the pea dehydrin sequence with sequences from other species revealed conserved amino acid regions: an N-terminal DEYGNP and a lysine-rich block (KIKEKLPG), both of which are present in two copies. Unexpectedly, pea dehydrin lacks a stretch of serine residues which is conserved in other dehydrins.
B12 mRNA and dehydrin proteins accumulated in dehydration-stressed seedlings, associated with elevated levels of endogenous abscisic acid (ABA). Applied ABA induced expression of dehydrins in unstressed seedlings. Dehydrin expression was rapidly reversed when seedlings were removed from the stress or from treatment with ABA and placed in water.
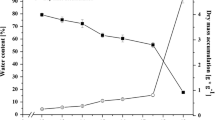
During pea cotyledon development, dehydrin mRNA and proteins accumulated in mid to late embryogenesis. Dehydrin proteins were some of the most actively synthesised at about the time of maximum fresh weight and represent about 2% of protein in mature cotyledons.
Similar content being viewed by others
References
Ariffin Z: Regulation of protein synthesis by ABA and PA in barley aleurone layers. M.Sc. Thesis, Australian National University, Canberra, Australia (1986).
Baker J, Steele C, Dure L: Sequence and characterization of 6 Lea proteins and their genes from cotton. Plant Mol Biol 11: 277–291 (1988).
Betlach MC, Hershfield V, Chow L, Brown W, Goodman HM, Boyer HW: A restriction endonuclease analysis of the bacterial plasmid controlling the Eco RI restriction and modification of DNA. Fedn Proc 35: 2037–2043 (1976).
Bradford KJ, Chandler PM: Expression of dehydrin-like proteins in embryos and seedlings of Zizania palustris and Oryza sativa during dehydration. Plant Physiol, in press (1992).
Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 (1976).
Brinckmann E, Hartung W, Wartinger M: Abscisic acid levels of individual leaf cells. Physiol Plant 80: 51–54 (1990).
Chandler PM, Higgins TJV, Randall PJ, Spencer D: Regulation of legumin levels in developing pea seeds under conditions of sulfur deficiency. Plant Physiol 71: 47–54 (1983).
Chandler PM, Spencer D, Randall PJ, Higgins TJV: Influence of sulfur nutrition on developmental patterns of some major pea seed proteins and their mRNAs. Plant Physiol 75: 651–657 (1984).
Chandler PM, Walker-Simmons M, King RW, Crouch ML, Close TJ: Expression of ABA-inducible genes in water-stressed cereal seedlings. J Cell Biol (Suppl 12C): 143 (1988).
Close TJ, Kort AA, Chandler PM: A cDNA-based comparison of dehydration-induced proteins (dehydrins) in barley and corn. Plant Mol Biol 13: 95–108 (1989).
Devereux J, Haeberli P, Smithies O: A comprehensive set of sequence analysis programs for the VAX. Nucl Acids Res 12: 387–395 (1984).
Galau GA, Close TJ: Sequences of the cotton group 2 LEA/RAB.dehydrin proteins encoded by Lea3 cDNAs. Plant Physiol, in press (1992).
Goday JA, Pardo JM, Pintor-Toro JA: A tomato cDNA inducible by salt stress and abscisic acid: nucleotide sequence and expression pattern. Plant Mol Biol 15: 695–705 (1990).
Goldberg RB, Barker SJ, Perez-Graul L: Regulation of gene expression during plant embryogenesis. Cell 56: 149–160 (1989).
Higgins TJB: Synthesis and regulation of major proteins in seeds. Anu Rev Plant Physiol 35: 191–221 (1984).
Hughes DW, Galau GA: Addition of the proteins to the cylindrical gel embedding medium for transverse molecular-weight markers in two-dimensional gel electrophoresis. Anal Biochem 140: 320–325 (1984).
Jacobsen JV, Chandler PM: Gibberellin and abscisic acid in germinating cereals. In: Davies PJ (ed), Plant Hormones and their Role in Plant Growth and Development, pp. 164–193. Martinus Nijhoff, Dordrecht (1987).
Jacobsen JV, Shaw DC: Heat-stable proteins and abscisic acid action in barley aleurone cells. Plant Physiol 91: 1520–1526 (1989).
Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 (1970).
Maniatis T, Fritsch EF, Sambrook L: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1982).
Millerd A, Spencer D: Changes in RNA-synthesizing activity and template activity in nuclei from cotyledons of developing pea seeds. Aust J Plant Physiol 1: 331–341 (1974).
Mundy J, Chua N-H: Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J 7: 2279–2286 (1988).
Piatkowski D, Schneider K, Salamini F, Bartels D: Characterization of five abscisic acid-responsive cDNA clones isolated from the desiccation tolerant plant Craterostigma plantagineum and their relationship to other water-stress genes. Plant Physiol 94: 1682–1688 (1990).
Pla M, Goday A, Vilardell J, Gómez J, Pagès M: Differential regulation of ABA-induced 23–25 kDa proteins in embryo and vegetative tissues of the viviparous mutants of maize. Plant Mol Biol 13: 385–394 (1989).
Proudfoot N: Poly(A) signals. Cell 64: 671–674 (1991).
Quatrano RS: Regulation of gene expression by abscisic acid during angiosperm embryo development. Oxford Surv Plant Mol Cell Biol 3: 467–476 (1986).
Raynal M, Gaubier P, Grellet F, Delseny M: Nucleotide sequence of a radish cDNA clone coding for a late embryogenesis abundant (LEA) protein. Nucl Acids Res 18: 6132 (1990).
Skriver K, Mundy J: Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2: 503–512 (1990).
Spencer D, Higgins TJV, Button SC, Davey RA: Pulse labelling studies on protein synthesis in developing pea seeds and evidence of a precursor form of legumin small subunit. Plant Physiol 66: 510–515 (1980).
Vilardell J, Goday A, Freire MA, Torent M, Martinez MC, Torne JM, Pages M: Gene sequence, developmental expression, and protein phosphorylation of RAB-17 in maize. Plant Mol Biol 14: 423–432 (1990).
Walker-Simmons M: ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol 84: 61–66 (1987).
Young RA, Davis RW: Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci USA 80: 1194–1198 (1983).
Zeevaart JAD, Creelman RA: Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol 39: 439–473 (1988).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Roberton, M., Chandler, P.M. Pea dehydrins: identification, characterisation and expression. Plant Mol Biol 19, 1031–1044 (1992). https://doi.org/10.1007/BF00040534
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00040534