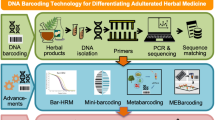
Abstract
Pharmacovigilance of herbal medicines relies on the product label information regarding the ingredients and the adherence to good manufacturing practices along the commercialisation chain. Several studies have shown that substitution of plant species occurs in herbal medicines, and this in turn poses a challenge to herbal pharmacovigilance as adverse reactions might be due to adulterated or added ingredients. Authentication of constituents in herbal medicines using analytical chemistry methods can help detect contaminants and toxins, but are often limited or incapable of detecting the source of the contamination. Recent developments in molecular plant identification using DNA sequence data enable accurate identification of plant species from herbal medicines using defined DNA markers. Identification of multiple constituent species from compound herbal medicines using amplicon metabarcoding enables verification of labelled ingredients and detection of substituted, adulterated and added species. DNA barcoding is proving to be a powerful method to assess species composition in herbal medicines and has the potential to be used as a standard method in herbal pharmacovigilance research of adverse reactions to specific products.
Similar content being viewed by others
Change history
02 January 2021
A Correction to this paper has been published: https://doi.org/10.1007/s40264-020-01029-9
References
Mann RD, Andrews EB. Pharmacovigilance. Chichester: Wiley; 2007.
World Health Organization. The importance of pharmacovigilance. Geneva: World Health Organization; 2002.
Meyboom RH, Egberts AC, Edwards IR, Hekster YA, de Koning FH, Gribnau FW. Principles of signal detection in pharmacovigilance. Drug Saf. 1997;16:355–65.
Bergvall T, Norén GN, Lindquist M. vigiGrade: a tool to identify well-documented individual case reports and highlight systematic data quality issues. Drug Saf. 2014;37:65–77.
Barnes J. Pharmacovigilance of herbal medicines. Drug Saf. 2003;26:829–51.
World Health Organization. WHO guidelines on safety monitoring of herbal medicines in pharmacovigilance systems. 2004. http://apps.who.int/iris/handle/10665/43034. Accessed 19 Jan 2015.
European Medicines Agency (EMA) Committee on Herbal Medicinal Products (HMPC). Guideline on quality of herbal medicinal products/traditional herbal medicinal products. London: EMA; 2011. Report no.: EMA/HMPC/201116/2005 Rev. 2. http://www.ema.europa.eu/ema/pages/includes/document/open_document.jsp?webContentId=WC500113209. Accessed 31 Jan 2015.
Applequist WL, Miller JS. Selection and authentication of botanical materials for the development of analytical methods. Anal Bioanal Chem. 2013;405:4419–28.
Pan S, Neeraj A, Srivastava KS, Kishore P, Danquah MK, Sarethy IP. A proposal for a quality system for herbal products. J Pharm Sci. 2013;102:4230–41.
Barnes J, Anderson LA, Phillipson JD. Herbal medicines: a guide for healthcare professionals. Pharmaceutical Press; 2003. http://www.cabdirect.org/abstracts/20033103050.html. Accessed 20 Jan 2015.
Wolfender J-L. HPLC in natural product analysis: the detection issue. Planta Med. 2009;75:719–34.
Li XQ, Sun XH, Cai S, Ying XX, Li FM. Investigation on the chemical constituents and variation of the flower buds of Lonicera species by UPLC-ESI-MS/MS and principle component analysis. Acta Pharm Sin. 2009;44:895–904.
Zhang C, Su J. Application of near infrared spectroscopy to the analysis and fast quality assessment of traditional Chinese medicinal products. Acta Pharm Sin B. 2014;4:182–92.
Jiang Y, David B, Tu P, Barbin Y. Recent analytical approaches in quality control of traditional Chinese medicines—a review. Anal Chim Acta. 2010;657:9–18.
Drašar P, Moravcova J. Recent advances in analysis of Chinese medical plants and traditional medicines. J Chromatogr B. 2004;812:3–21.
Scora RW. Problems in chemotaxonomy: the influence of varying soil conditions, of geographical and individual variants upon the distribution of certain substances in chromatographed extracts of Monarda fistulosa. Plant Soil. 1966;24:145–52.
Zewdie Y, Bosland PW. Capsaicinoid profiles are not good chemotaxonomic indicators for Capsicum species. Biochem Syst Ecol. 2001;29:161–9.
Hebert PDN, Cywinska A, Ball S, de Waard J. Biological identifications through DNA barcodes. Proc R Soc B. 2003;270:313–22.
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA. 2005;102:8369–74.
Jarman SN, Elliott NG. DNA evidence for morphological and cryptic Cenozoic speciations in the Anaspididae’, living fossils’ from the Triassic. J Evol Biol. 2000;13:624–33.
Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proc Natl Acad Sci USA. 2008;105:6350–5.
Ragupathy S, Newmaster SG, Murugesan M, Balasubramaniam V. DNA barcoding discriminates a new cryptic grass species revealed in an ethnobotany study by the hill tribes of the Western Ghats in southern India. Mol Ecol Resour. 2009;9:164–71.
Liu JIE, Moeller M, Gao L-M, Zhang D-Q, Li D-Z. DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol Ecol Resour. 2011;11:89–100.
De Carvalho MR, Bockmann FA, Amorim DS, Brandão CRF, de Vivo M, de Figueiredo JL, et al. Taxonomic impediment or impediment to taxonomy? A commentary on systematics and the cybertaxonomic-automation paradigm. Evol Biol. 2007;34:140–3.
Godfray HCJ. Linnaeus in the information age. Nature. 2007;446:259–60.
Boessenkool S, McGlynn G, Epp LS, Taylor D, Pimentel M, Gizaw A, et al. Use of ancient sedimentary DNA as a novel conservation tool for high-altitude tropical biodiversity. Conserv Biol. 2014;28:446–55.
Parducci L, Jørgensen T, Tollefsrud MM, Elverland E, Alm T, Fontana SL, et al. Glacial survival of boreal trees in Northern Scandinavia. Science. 2012;335:1083–6.
Willerslev E, Davison J, Moora M, Zobel M, Coissac E, Edwards ME, et al. Fifty thousand years of Arctic vegetation and megafaunal diet. Nature. 2014;506:47–51.
Van Geel B, Aptroot A, Baittinger C, Birks HH, Bull ID, Cross HB, et al. The ecological implications of a Yakutian mammoth’s last meal. Q Res. 2008;69:361–76.
Coghlan M, Haile J, Houston J, Murray D, White N, Moolhuijzen P, et al. Deep sequencing of plant and animal DNA contained within traditional chinese medicines reveals legality issues and health safety concerns. PLoS Genet. 2012;8:e1002657.
Newmaster SG, Grguric M, Shanmughanandhan D, Ramalingam S, Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11:222.
Kerr KCR, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, Hebert PDN. Comprehensive DNA barcode coverage of North American birds. Mol Ecol Notes. 2007;7:535–43.
Smith MA, Poyarkov NA, Hebert PDN. CO1 DNA barcoding amphibians: take the chance, meet the challenge. Mol Ecol Resour. 2008;8:235–46.
Eurlings MCM, Heuveling van Beek H, Gravendeel B. Polymorphic microsatellites for forensic identification of agarwood (Aquilaria crassna). Forensic Sci Int. 2010;197:30–4.
Chen S, Yao H, Han J, Liu C, Song J, Shi L, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:1–8.
Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA. 2011;108:19641–6.
Kool A, de Boer HJ, Krüger Å, Rydberg A, Abbad A, Björk L, et al. Molecular identification of commercialized medicinal plants in Southern Morocco. PLoS One. 2012;7:e39459.
Thompson KA, Newmaster SG. Molecular taxonomic tools provide more accurate estimates of species richness at less cost than traditional morphology-based taxonomic practices in a vegetation survey. Biodivers Conserv. 2014;23:1411–24.
Cho Y, Mower JP, Qiu Y, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci USA. 2004;101:17741–6.
Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007;2:e508.
Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One. 2008;3:e2802.
Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, et al. Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol Ecol Resour. 2009;9:130–9.
Ford CS, Ayres KL, Toomey N, Haider N, van Alphen Stahl J, Kelly LJ, et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc. 2009;159:1–11.
CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106:12794–7.
Hollingsworth PM. Refining the DNA barcode for land plants. Proc Natl Acad Sci USA. 2011;108:19451–2.
Burgess KS, Fazekas AJ, Kesanakurti PR, Graham SW, Husband BC, Newmaster SG, et al. Discriminating plant species in a local temperate flora using the rbcL + matK DNA barcode. Methods Ecol Evol. 2011;2:333–40.
Paton AJ, Brummitt N, Govaerts R, Harman K, Hinchcliffe S, Allkin B, et al. Towards Target 1 of the Global Strategy for Plant Conservation: a working list of all known plant species—progress and prospects. Taxon. 2008;57:602–11.
Joppa LN, Roberts DL, Pimm SL. How many species of flowering plants are there? Proc Biol Sci. 2011;278(1705):554–9.
Newmaster SG, Fazekas AJ, Ragupathy S. DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Botany. 2006;84:335–41.
De Vere N, Rich TC, Ford CR, Trinder SA, Long C, Moore CW, et al. DNA barcoding the native flowering plants and conifers of Wales. PLoS One. 2012;7:e37945.
Kuzmina ML, Johnson KL, Barron HR, Hebert PD. Identification of the vascular plants of Churchill, Manitoba, using a DNA barcode library. BMC Ecol. 2012;12:25.
Liu J, Yan H-F, Newmaster SG, Pei N, Ragupathy S, Ge X-J. The use of DNA barcoding as a tool for the conservation biogeography of subtropical forests in China. Divers Distrib. 2015;21:188–99.
Ebihara A, Nitta JH, Ito M. Molecular species identification with rich floristic sampling: DNA barcoding the pteridophyte flora of Japan. PLoS One. 2010;5:e15136.
De Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–86.
Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–88.
Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, et al. Recently formed polyploid plants diversify at lower rates. Science. 2011;333:1257.
Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106:13875–9.
Knowles LL, Carstens BC. Delimiting species without monophyletic gene trees. Syst Biol. 2007;56:887–95.
De Boer HJ, Ouarghidi A, Martin G, Abbad A, Kool A. DNA barcoding reveals limited accuracy of identifications based on folk taxonomy. PLoS One. 2014;9:e84291.
Ratnasingham S, Hebert PD. BOLD: The barcode of life data system. Mol Ecol Notes. 2007;7:355–64. http://www.barcodinglife.org. Accessed 31 Jan 2015.
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42.
Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Spouge JL, Mariño-Ramírez L. The practical evaluation of DNA barcode efficacy. Methods Mol Biol. 2012;858:365–77.
Hinchliff CE, Smith SA. Some limitations of public sequence data for phylogenetic inference (in plants). PLoS One. 2014;9:e98986.
Lou S-K, Wong K-L, Li M, But PP, Tsui SK, Shaw P-C. An integrated web medicinal materials DNA database: MMDBD (Medicinal Materials DNA Barcode Database). BMC Genom. 2010;11:402.
Heubl G. New aspects of DNA-based authentication of Chinese medicinal plants by molecular biological techniques. Planta Med. 2010;76:1963–74.
Techen N, Parveen I, Pan Z, Khan IA. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 2014;25:103–10.
Purushothaman N, Newmaster SG, Ragupathy S, Stalin N, Suresh D, Arunraj DR, et al. A tiered barcode authentication tool to differentiate medicinal Cassia species in India. Genet Mol Res. 2014;13:2959–68.
Little DP, Gulick P. Authentication of Ginkgo biloba herbal dietary supplements using DNA barcoding. Genome. 2014;57:1–4.
Kazi T, Hussain N, Bremner P, Slater A, Howard C. The application of a DNA-based identification technique to over-the-counter herbal medicines. Fitoterapia. 2013;87:27–30.
Wallace LJ, Boilard SM, Eagle SH, Spall JL, Shokralla S, Hajibabaei M. DNA barcodes for everyday life: routine authentication of natural health products. Food Res Int. 2012;49:446–52.
Zuo Y, Chen Z, Kondo K, Funamoto T, Wen J, Zhou S. DNA barcoding of Panax species. Planta Med. 2011;77:182–7.
Vassou SL, Kusuma G, Madasamy P. DNA barcoding for species identification from dried and powdered plant parts: a case study with authentication of the raw drug market samples of Sida cordifolia. Gene. 2015;559(1):86–93.
Seethapathy GS, Ganesh D, Kumar JUS, Senthilkumar U, Newmaster SG, Ragupathy S, et al. Assessing product adulteration in natural health products for laxative yielding plants, Cassia, Senna, and Chamaecrista, in Southern India using DNA barcoding. Int J Legal Med. 2014;1–8.
Baker DA. DNA barcode identification of black cohosh herbal dietary supplements. J AOAC Int. 2012;95:1023–34.
Stoeckle MY, Gamble CC, Kirpekar R, Young G, Ahmed S, Little DP. Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci Rep. 2011;1:1–7.
Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol. 2012;21:2045–50.
Jørgensen T, Kjaer KH, Haile J, Rasmussen M, Boessenkool S, Andersen K, et al. Islands in the ice: detecting past vegetation on Greenlandic nunataks using historical records and sedimentary ancient DNA Meta-barcoding. Mol Ecol. 2012;21:1980–8.
Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac E, Pompanon F, et al. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol Resour. 2009;9:51–60.
Veldman S, Otieno JN, van Andel T, Gravendeel B, de Boer HJ. Efforts urged to tackle thriving illegal orchid trade in Tanzania and Zambia for chikanda production. Traffic Bull. 2014;26:47–50.
Reja V, Kwok A, Stone G, Yang L, Missel A, Menzel C, et al. ScreenClust: advanced statistical software for supervised and unsupervised high resolution melting (HRM) analysis. Methods. 2010;50:S10–4.
Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–60.
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–60.
Osathanunkul M, Madesis P, de Boer HJ. Bar-HRM for authentication of plant-based medicines: evaluation of three medicinal products derived from Acanthaceae species. PLoS One. 2015;10(5):e0128476.
Madesis P, Ganopoulos I, Sakaridis I, Argiriou A, Tsaftaris A. Advances of DNA-based methods for tracing the botanical origin of food products. Food Res Int. 2014;60:163–72.
Ganopoulos I, Madesis P, Darzentas N, Argiriou A, Tsaftaris A. Barcode High Resolution Melting (Bar-HRM) analysis for detection and quantification of PDO “Fava Santorinis” (Lathyrus clymenum) adulterants. Food Chem. 2012;133:505–12.
Ganopoulos I, Bazakos C, Madesis P, Kalaitzis P, Tsaftaris A. Barcode DNA high-resolution melting (Bar-HRM) analysis as a novel close-tubed and accurate tool for olive oil forensic use. J Sci Food Agric. 2013;93:2281–6.
Ganopoulos I, Xanthopoulou A, Mastrogianni A, Drouzas A, Kalivas A, Bletsos F, et al. High resolution melting (HRM) analysis in eggplant (Solanum melongena L.): a tool for microsatellite genotyping and molecular characterization of a Greek Genebank collection. Biochem Syst Ecol. 2015;58:64–71.
Kalivas A, Ganopoulos I, Xanthopoulou A, Chatzopoulou P, Tsaftaris A, Madesis P. DNA barcode ITS2 coupled with high resolution melting (HRM) analysis for taxonomic identification of Sideritis species growing in Greece. Mol Biol Rep. 2014;41:5147–55.
Schmiderer C, Mader E, Novak J. DNA-based identification of Helleborus niger by high-resolution melting analysis. Planta Med. 2010;76:1934–7.
Ganopoulos I, Aravanopoulos F, Madesis P, Pasentsis K, Bosmali I, Ouzounis C, et al. Taxonomic identification of Mediterranean pines and their hybrids based on the high resolution melting (HRM) and trnL approaches: from cytoplasmic inheritance to timber tracing. PLoS One. 2013;8:e60945.
Madesis P, Ganopoulos I, Bosmali I, Tsaftaris A. Barcode High Resolution Melting analysis for forensic uses in nuts: a case study on allergenic hazelnuts (Corylus avellana). Food Res Int. 2013;50:351–60.
Madesis P, Ganopoulos I, Anagnostis A, Tsaftaris A. The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control. 2012;25:576–82.
Bosmali I, Ganopoulos I, Madesis P, Tsaftaris A. Microsatellite and DNA-barcode regions typing combined with High Resolution Melting (HRM) analysis for food forensic uses: a case study on lentils (Lens culinaris). Food Res Int. 2012;46:141–7.
Ganopoulos I, Madesis P, Tsaftaris A. Universal ITS2 barcoding DNA region coupled with high-resolution melting (HRM) analysis for seed authentication and adulteration testing in leguminous forage and pasture species. Plant Mol Biol Rep. 2012;30:1322–8.
Lai G-H, Chao J, Lin M-K, Chang W-T, Peng W-H, Sun F-C, et al. Rapid and sensitive identification of the herbal tea ingredient Taraxacum formosanum using loop-mediated isothermal amplification. Int J Mol Sci. 2015;16:1562–75.
Novak J, Grausgruber-Gröger S, Lukas B. DNA-based authentication of plant extracts. Food Res Int. 2007;40:388–92.
Smyth RP, Schlub TE, Grimm A, Venturi V, Chopra A, Mallal S, et al. Reducing chimera formation during PCR amplification to ensure accurate genotyping. Gene. 2010;469:45–51.
Lammers Y, Peelen T, Vos RA, Gravendeel B. The HTS barcode checker pipeline, a tool for automated detection of illegally traded species from high-throughput sequencing data. BMC Bioinform. 2014;15:44.
Food and Drug Administration, HHS. Current good manufacturing practice in manufacturing, packaging, labeling, or holding operations for dietary supplements. Final rule. Fed Regist. 2007;72(121):34751–958 http://www.ncbi.nlm.nih.gov/pubmed/17674484. Accessed 24 April 2015.
FDA. FDA response to Heinrich and Hatch. Silver Spring: FDA; 2015. http://www.npainfo.org/App_Themes/NPA/docs/regulatoryLegislative/FDA/FDA%20Response_Hatch-Heinrich%20Letter_Dietary%20Supple-ments.pdf. Accessed 18 Apr 2015.
Palhares RM, Gonçalves Drummond M, dos Santos Alves Figueiredo Brasil B, Pereira Cosenza G, das Graças Lins Brandão M, Oliveira G. Medicinal plants recommended by the World Health Organization: DNA barcode identification associated with chemical analyses guarantees their quality. PLoS ONE 2015;10(5):e0127866. doi:10.1371/journal.pone.0127866.
Acknowledgments
The research leading to these results has received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007–2013/under REA (Research Executive Agency) Grant Agreement No. PITN-GA-2013-606895 (to HdB), the Norway-Romania EEA (European Economic Area) Research Programme operated by the MECS-ANCSI PO (Ministerul Educatiei si Cercetarii Stiintifice - Autoritatea Nationala pentru Cercetare Stiintifica si Inovare Programme Operator) under the EEA Financial Mechanism 2009–2014 Project Contract No. 2SEE/2014 (to HdB and MI), the Core Program PN09-360402/BIODIV (MECS-ANCSI) (to MI) and the International Science and Technology Partnership Canada and the Ontario Ministry of Economic Development, Trade and Employment (MEDI) (to SN).
Conflict of interest
Hugo de Boer, Mihael Ichim and Steven Newmaster have no conflicts of interest that are directly relevant to the content of this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Boer, H.J., Ichim, M.C. & Newmaster, S.G. DNA Barcoding and Pharmacovigilance of Herbal Medicines. Drug Saf 38, 611–620 (2015). https://doi.org/10.1007/s40264-015-0306-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-015-0306-8