Abstract
The complex nature of familial cardiomyopathies is incompletely understood. The effective management of patients and their relatives therefore requires a multidisciplinary approach involving a specialist genetic counselling service. Genetic testing is clinically available, and our knowledge in this area continues to grow with developments in new technologies. Many of the genes associated are not specific to a particular type of cardiomyopathy, and recent data suggests that some patients may have more than one mutation or variant contributing to disease. Developments in next generation sequencing have enabled us to accurately and efficiently sequence these areas of interest, but the current capacity to analyse and interpret this data (bioinformatics) remains a major limitation. Genetic guided therapies have the potential to revolutionise our management of affected patients in the future but this is far from being a clinical reality at the current time.
Similar content being viewed by others
Introduction
Cardiomyopathies are a heterogeneous group of diseases of the myocardium associated with ventricular and atrial arrhythmias, cardiac failure, stroke and sudden death [1]. The first causative genes were identified in 1990 [2, 3] and were found to be associated with hypertrophic cardiomyopathy (HCM) which is the most common form, with an estimated prevalence of 1 in 500 in Europe. It is characterised by unexplained cardiac hypertrophy accompanied by myocyte disarray and myocardial fibrosis. Arrhythmogenic right ventricular cardiomyopathy (ARVC) and familial idiopathic dilated cardiomyopathy (DCM) are less common. Patients with ARVC may have relatively preserved cardiac morphology with a propensity towards ventricular arrhythmias, or have progressive disease characterised by myocyte loss, inflammation and fibrofatty deposition that predominates in the right ventricle, but may also involve the left ventricle. DCM on the other hand, is characterised by systolic dysfunction and left ventricular dilatation. Other rarer inherited forms include left ventricular non-compaction cardiomyopathy (LVNC) and restrictive cardiomyopathy (RCM). Collectively, heritable cardiomyopathies affect about 1 in 390 people [1], but this may be an underestimate of the true prevalence, given the incomplete or age-related penetrance of disease and lack of symptoms amongst most carriers.
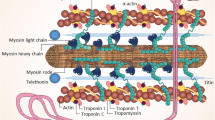
Most identified mutations associated with cardiomyopathy are inherited in an autosomal dominant manner. The exceptions include rare autosomal recessive and X-linked disease, as well as maternally inherited mitochondrial cardiomyopathy, which is usually associated with multisystem disease and presents in infancy. Table 1 provides an overview of the genes that have been shown to be associated with the three most prevalent inherited cardiomyopathies. There is huge genetic heterogeneity and genetic overlap in these conditions. For example, mutations in the myosin heavy chains, myosin-binding protein C, cardiac troponins and calcium handling protein genes are associated with more than one type of cardiomyopathy. DCM in particular, has been associated with a large number different genes [8]. Moreover, within the individual genes, multiple mutations and variants have been described and novel mutations private to individual families continue to be identified. Translating this vast and complex science into clinical practice has been challenging. Despite the large number of genes known to be associated with cardiomyopathy, a clear genetic cause has only been identified in up to 65 % of familial HCM cases, and this figure is even lower for ARVC and DCM, being quoted as 50 % and 30–35 % respectively [9].
Current Utility of Gene Testing in Cardiomyopathies
The Heart Rhythm Society/European Heart Association (HRS/EHRA) international consensus statement published in 2011 [10••] summarises the current evidence and expert opinion on the utility of genetic testing for heritable cardiomyopathies and the potential diagnostic, prognostic and therapeutic impact of a genetic test result in such circumstances. As with any test, it is recommended that treatment decisions should not solely rely on a patient’s genetic test result, but should be based on an individual’s comprehensive clinical evaluation. They identify a substantial knowledge gradient among cardiologists regarding inherited cardiomyopathies, highlighting the importance of managing affected patients and families at specialist centres equipped with the expertise and facilities to provide counselling, carry out genetic evaluation, and effectively manage affected patients and family members [10••].
It is currently not possible to completely exclude familial cardiomyopathy by genetic testing alone, unless a specific disease-causing mutation has been identified in another member of the family. In the case of HCM, comprehensive or targeted (MYBPC3, MYH7, TNNI3, TNNT2, TPM1) testing is recommended in the proband if there is a firm diagnosis of HCM, followed by cascade genetic screening of the identified mutation in the family [10••]. Evidence supports starting clinical and genetic screening of first-degree relatives of patients affected with HCM from 10 years of age onwards, and including such relatives in screening until at least the age of 60 years. It has been suggested that screening of more distant relatives is most effective in the early adolescent and teenage years [11•]. This strategy may also be cost-effective in the long term by focusing clinical resources [12, 13•].
In the case of ARVC, the HRS/EHRA consensus panel advises that comprehensive or targeted testing for DSC2, DSG2, DSP, JUP, PKP2, and TMEM43 mutations can be useful for patients with a clear diagnosis of ARVC that satisfies the 2010 taskforce criteria [10••, 14]. It is unlikely to be helpful however, if the diagnosis is borderline. Mutation-specific genetic testing is then recommended for family members following identification of a causative mutation in the proband. The guidelines are similar for DCM, LVNC and RCM except that even less is known about the genes causative of RCM, and genetic testing may be considered for patients with a high suspicion of disease but is likely to be less useful. Nonetheless, cascade testing in the family is still recommended if a causative gene is found in the proband.
Mutation-specific genetic testing amongst members of families with a positive cardiomyopathy gene test may have diagnostic and therefore management implications. Clinicians need to beware of potential age-related penetrance in asymptomatic or clinically unaffected mutation carriers who therefore require follow-up. However, asymptomatic mutation negative relatives can potentially be discharged from follow up. It should also be noted that at present there is limited prognostic role of genetic testing except for the increased risk of sudden cardiac death in lamin A/C and desmosome-mediated DCM [10••].
Genetic tests are probabilistic rather than deterministic, and as highlighted above, the yield from genetic testing is far from perfect in the current context. It is therefore recommended that physicians managing a patient with genotype-negative disease liaise with specialist genetic research laboratories to identify novel, potentially disease-causing mutations [10••, 15]. As more novel genes are added to the available genetic test panels, more mutations and variants will be identified, but the test specificity will inevitably decrease until more is understood about disease causation. Appropriate counselling and a multidisciplinary approach involving geneticists and/or a genetic counselling service are vital to prepare patients and family members adequately before, during and after the screening process. The psychological, economic and social impact of genetic diagnosis in asymptomatic mutation carriers should also be carefully considered. For example, making a genetic diagnosis in an asymptomatic athlete with a family history of cardiomyopathy may have major implications on their career [16, 17].
As our understanding of the genetics of inherited cardiomyopathies improves, there will be greater utilisation of pre-implantation genetic testing in at-risk families in the context of assisted reproduction. It is important to consider the potential role of such testing in the clinical setting and the practical, ethical, social and cultural issues thus raised. Some centres already offer pre-implantation genetic diagnosis for cardiomyopathies based on our existing knowledge of causative mutations, and this area is likely to expand in the future. As with all genetic testing, specialist facilities for pre- and post-test genetic counselling including discussion of the limits in our knowledge of these conditions, risks of testing, benefits and options available should be an integral part of management [10••, 18].
Recent Developments in Diagnostics and Gene Discovery
Newer next-generation sequencing (NGS) methods have resulted in rapid advances in our understanding of the heritable cardiomyopathies and their complex genetic associations.
Targeted sequencing of the exome (the protein coding region of the genome) is currently being used in diagnostics. However many of the causative or associated genes in familial cardiomyopathies are yet to be identified, especially in DCM. Using targeted sequencing therefore reduces the diagnostic yield. Although cardiomyopathy panels with genes known to have causative association are being developed for clinical use, these may become outdated as newer genes are discovered [9]. ‘Crossover phenotypes’ add to the complexity of genetic diagnosis because many of the genes responsible for inherited cardiomyopathies are not specific to a particular phenotype. For example, desmosomal protein gene mutations have been more typically associated with ARVC, but only 30–50 % of patients with ARVC have mutations in these genes [19]. Interpretation of desmosomal mutations is complicated by their presence in patients with a diagnosis idiopathic DCM and also by the high prevalence of variants of these genes in the general population [20•, 21].
In 2011, Meder and colleagues presented an approach to carry out comprehensive genetic screening in patients with hereditary DCM or HCM in a fast and cost-efficient manner using high-throughput mutation screening with microarray-based target enrichment followed by a next-generation sequencing platform (SOLiD™ NGS) [22•]. This enabled accurate detection of sequence variants and mutations in multiple genetic loci of interest in a time- and cost-efficient manner. Although the costs of NGS are still substantial in terms of investment in equipment, the cost per base is markedly lower than for traditional ‘Sanger’ sequencing. Moreover, use of parallel technology as in SOLiD™ NGS reduces manual handling time and can cost substantially less than Sanger sequencing alone with results available within 2 weeks. Proving disease causality is, however, key to clinical genetic diagnostics. An initial approach is the comparison of detected variants against databases of gene mutations that have already been shown to be causative [22•]. This, however, relies on the stringency of previous reports and limits interpretation of novel mutations. Further evaluation of the mutation can be achieved by cosegregation of phenotype with genotype in a family provided the pedigree is sufficiently large and accessible [23]. Integration with data from in silico tools and functional studies will be vital in many cases.
One example of the potential success of new technologies is TTN, the largest gene in the genome and a known cause of DCM. TTN encodes the sarcomeric protein titin and large truncating mutations have been observed recently in approximately 25 % of cases of familial DCM probands making it the most important single source of genetic variation in the condition [24•]. Due to the size of the gene, these could only be detected practically using NGS technologies. Although the functional role of titin in the cardiomyocyte are known, the effects of TTN truncating mutations on the exact pathophysiology of DCM have yet to be defined. To complicate matters further, titin mutations have also recently been associated with ARVC [25]. Much work therefore remains to analyse and validate these exciting genetic data before they can be applied clinically.
One approach to discovery of new genes involved in disease is whole exome or genome sequencing (WGS) using NGS. This has the advantage of sequencing all relevant genetic variation but at the expense of generating massive datasets that require extensive input from bioinformaticians for analysis. For example, a missense mutation in MRPL3, which encodes a large mitochondrial ribosomal protein, was recently identified in a family with HCM using exome sequencing [26]. In this study, the mutation was shown to alter the stability of MRPL3 and was associated with defective assembly of the ribosomal subunit. In another study, a homozygous missense mutation in AARS2, which was later shown to encode mitochondrial alanyl-tRNA synthetase (mtAlaRS), was identified through exome sequencing in early infantile DCM [27]. These cases highlight the potential clinical utility of exome sequencing and similar studies on non-mitochondrial genes are crucial to novel gene discovery.
Genome-wide association studies utilising a single nucleotide polymorphism (SNP) array have identified copy number variants associated with sporadic cardiomyopathies. This method associated DCM with the heat shock protein cochaperone BCL2-associated athanogene 3 (BAG3). Subsequent exome sequencing enabled identification of mutations and variants in familial cases that segregated with disease [28, 29]. Genome-wide mapping involving linkage analysis and exome sequencing has also been used to identify GATAD1 as a gene associated with autosomal recessive DCM [30].
A study that used multiplex ligation-depended probe amplification analysis to characterise pathogenic PKP2 mutations underlying ARVC has raised a note of caution about the role of NGS [31•]. This successfully identified large deletions in PKP2, which may have otherwise been missed by NGS and illustrated a potential limitation of the technology.
The Need for Functional Characterisation
Increasing feasibility, efficiency and reducing costs of NGS technologies will result in more comprehensive sequencing of genes in affected patients. This will lead inevitably to the identification of numerous genetic variants of uncertain significance, as well as novel genetic loci. It will be hard to justify the use of this expanding knowledge base in clinical practice without robust data on genotype–phenotype correlation or functional assays. It is therefore crucially important to determine the extent of natural genetic variation and to distinguish ‘background noise’, particularly on a population and ethnicity basis, to define how much is disease-associated and therefore disease-modifying and/or disease-causative. For example, a study of general population exome data from the National Heart, Lung, and Blood Institute’s Exome Sequencing Project has demonstrated that previously disease-associated mutations in the HCM genes were present in a higher frequency than expected for a purely monogenic disease model [32].
More research and investment is needed in the development of efficient, cost-effective functional assays to answer these questions and to determine the significance of the newer variants being discovered. Zebrafish knockouts would be one potentially useful model for high throughput work for further assessing these areas of interest, but the obvious limitations include the physiological differences between the zebrafish models and the human heart [33]. Zebrafish models have, however, been used to demonstrate the role of BAG3 mutations in DCM [28]. Another alternative is to use mouse models [34, 35]. A recent functional study in mice demonstrated that certain mutations in MYPN may result in disturbed myofibrillogenesis or disrupted intercalated discs leading to various forms of cardiomyopathy, including HCM and RCM [35]. Another transgenic mouse model has been used to demonstrate the role of microRNAs in regulating post-transcriptional gene expression in early and end-stage HCM [36]. Such studies are important in providing functional evidence for the pathophysiological mechanisms of disease, but are much more expensive and less feasible for high throughput work than the zebrafish models.
In silico models backed up by in vitro and in vivo data may be used to obtain further data on the functional effects of the mutations and variants of interest but their reliability has been unclear [37]. Novel computational methods have been developed that may potentially contribute to the classification and aid in the analysis of the predictive function of these variants in HCM in particular [38].
Currently, however, the speed at which NGS can identify variants of uncertain significance is far greater than the speed at which functional assays can be used to determine variant pathogenicity. A recent study evaluated the extent of genetic variation amongst the ARVC susceptibility genes PKP2 (plakophilin), DSP (desmoplakin), DSG2 (desmoglein 2), DSC2 (desmocollin 2) and TMEM43 (transmembrane protein 43) in a sample of over 200 healthy subjects from the general population [39••]. It was concluded that whilst mutations causing severe impact on protein structure (so-called radical mutations) are associated with a high probability of developing ARVC, missense mutations were common and should be interpreted with caution in the context of race, ethnicity, location of the mutation and sequence conservation across species.
Developments in Genotype Phenotype Correlation
Caution must be exercised when interpreting small cohort studies with regard to genotype–phenotype correlation. For example, studies have suggested that mutations in the cardiac myosin binding protein-C (MYBPC3) may only cause late-onset benign HCM. A subsequent analysis on a large series of families with HCM and underlying MYBPC3 mutations showed that there is marked heterogeneity in disease expression with incomplete age-related and gender-specific penetrance [40]. Hence, caution must be exercised when carrying out cascade screening and risk assessment in family members of affected probands based on the current level of evidence on known mutations and our limited knowledge on newer mutations. Data from another observational study indicates that the presence of more than one sarcomeric mutation (known as compound heterozygosity or polygenic disease) may confer a predisposition to adverse disease progression in affected patients [41•]. These data support a polygenic inheritance in some forms of cardiomyopathy. Compound heterozygosity and polygenicity has also been demonstrated in ARVC [42, 43]. Quarta et al. found that relatives harbouring more than one genetic variant had a significantly increased risk of developing ARVC, an important factor when interpreting the variable expression of disease within families. This study also highlighted that penetrance may be definition-dependent, being greater with the 2010 task force criteria compared to the 1994 criteria in the case of ARVC [44].
Some mutations have been found to confer a higher susceptibility to malignant arrhythmias. Lamin A/C (LMNA) mutations have classically been associated with DCM and have an important role in risk stratifying those DCM patients who are at high risk of malignant ventricular arrhythmias [45]. A recent study by Quarta et al. [46•] also found lamin A/C mutations in severe forms of ARVC. It was recommended by the investigators that lamin A/C should be added to a desmosomal gene panel when genetically testing for suspected ARVC. Similar observations have been made with phospholamban mutations, which are implicated in both DCM and ARVC. A recent study showed that PLN R14del+ patients diagnosed with DCM were more prone to an arrhythmogenic phonotype [47]. This supports the concept of ‘arrhythmogenic cardiomyopathy’ with a common underlying mutation in the Phospholamban gene amongst patients traditionally diagnosed as DCM or ARVC. Plakophilin 2 (PKP2) mutations, which have previously been associated with ARVC, too have recently been demonstrated in post mortem cardiac tissue sample in cases of sudden death with negative autopsy [48]. These data suggest an association between genetic variation in desmosomal genes and fatal arrhythmias without histopathological evidence of disease at autopsy.
Therapeutic Implications of Genetic Testing
The current therapeutic utility of genetic testing is limited to identification of mutations that may be associated with a high rate of ventricular arrhythmias or sudden cardiac death, and hence as a risk stratification tool to aid decision making about prophylactic ICD (implantable cardioverter defibrillator) therapy in affected patients. As described above, DCM patients with LMNA mutations have a higher propensity to develop prominent cardiac conduction system disease and pre-emptive use of ICD therapy has been advocated prior to the occurrence of significant symptoms [5].
It is hoped that developments in our knowledge of this area may enable genetically directed prophylactic pharmacotherapy in the future. In a minority of patients who have cardiomyopathy as part of a more global multisystem disease such as Fabry disease, genetic test results already have direct therapeutic implications in terms of enzyme replacement [10••]. In mutations of the X-linked lysosome-associated membrane protein gene, LAMP2, early molecular diagnosis and timely cardiac transplantation has been shown to be associated with better clinical outcome [49]. The immunosuppressant rapamycin (sirolimus) has been suggested as a potential new therapy in attenuating cardiomyopathy. Recent genetic evidence from its use in Zebrafish and rodent models suggest that TOR (the mammalian target of rapamycin) signalling inhibition may present a novel therapeutic strategy in the management of cardiomyopathy [50–52].
Conclusions
Our understanding of the genetics underlying heritable cardiomyopathies is rapidly evolving, and it is likely to change further over the next few years with advances in NGS methods and bioinformatics. These new technologies have enabled the discovery of novel genes associated with disease, but there exists a major gap in knowledge when assigning clinical relevance to new mutations and variants of uncertain significance. A single phenotype may be accounted for by a multitude of genes and mutations in each individual gene may give rise to different phenotypes in different people. This vast heterogeneity makes research in this area fascinating, but makes clinical application difficult. One of the main challenges over the next few years would be in designing cost-effective and time-efficient functional studies to demonstrate the effects of new variants on pathophysiology of disease. It is imperative that clinicians managing affected patients and their families are not only aware of new developments but also of the significance of these new developments in the context of their patients.
Abbreviations
- HCM:
-
Hypertrophic cardiomyopathy
- DCM:
-
Dilated cardiomyopathy
- ARVC:
-
Arrhythmogenic right ventricular cardiomyopathy
- RCM:
-
Restrictive cardiomyopathy
- LVNC:
-
Left ventricular non-compaction cardiomyopathy
- NGS:
-
Next generation sequencing
- WGS:
-
Whole genome sequencing
- ESP:
-
Exome Sequencing Project
- MLPA:
-
Multiplex ligation-depended probe amplification
- SNP:
-
Single nucleotide polymorphism
- ICD:
-
Implantable cardioverter defibrillator
- HRS/EHRA:
-
Heart Rhythm Society/European Heart Association
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Raju H, Alberg C, Sagoo GS. Inherited cardiomyopathies. BMJ. 2011;343:d6966.
Tanigawa G, Jarcho JA, Kass S, et al. A molecular basis for familial hypertrophic cardiomyopathy: an alpha/beta cardiac myosin heavy chain hybrid gene. Cell. 1990;62(5):991–8.
Geisterfer-Lowrance AA, Kass S, Tanigawa G, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62(5):999–1006.
Bos JM, Ommen SR, Ackerman MJ. Genetics of hypertrophic cardiomyopathy: one, two, or more diseases? Curr Opin Cardiol. 2007;22(3):193–9.
Hershberger RE, Lindenfeld J, Mestroni L, et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J Cardiac Fail. 2009;15(2):83–97.
Hershberger RE, Siegfried JD. Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2011;57(16):1641–9.
Parvari R, Levitas A. The mutations associated with dilated cardiomyopathy. Biochem Res Int. 2012;2012:639250.
Lakdawala NK, Funke BH, Baxter S, et al. Genetic testing for dilated cardiomyopathy in clinical practice. J Cardiac Fail. 2012;18(4):296–303.
Norton N, Li D, Hershberger RE. Next-generation sequencing to identify genetic causes of cardiomyopathies. Curr Opin Cardiol. 2012;27(3):214–20.
•• Ackerman MJ, Priori SG, Willems S, et al.: HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm. 2011;8(8):1308–39.
Expert consensus document with current guidelines for genetic testing in clinical practice
• Nannenberg EA, Michels M, Christiaans I, et al.: Mortality risk of untreated myosin-binding protein C-related hypertrophic cardiomyopathy: insight into the natural history. J Am Coll Cardiol. 2011;58(23):2406–14.
Provides evidence on the prognostic implications of MYBPC mutations
Wordsworth S, Leal J, Blair E, et al. DNA testing for hypertrophic cardiomyopathy: a cost-effectiveness model. Eur Heart J. 2010;31(8):926–35.
• Ingles J, McGaughran J, Scuffham PA, et al.: A cost-effectiveness model of genetic testing for the evaluation of families with hypertrophic cardiomyopathy. Heart. 2012;98(8):625–30.
Provides evidence on cost-effective genetic testing in the clinical setting
Marcus FI, McKenna WJ, Sherrill D, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J. 2010;31(7):806–14.
Judge DP. Arrhythmogenic right ventricular dysplasia/cardiomyopathy: a family affair. Circulation. 2011;123(23):2661–3.
Borjesson M, Serratosa L, Carre F, et al. Consensus document regarding cardiovascular safety at sports arenas: position stand from the European Association of Cardiovascular Prevention and Rehabilitation (EACPR), section of Sports Cardiology. Eur Heart J. 2011;32(17):2119–24.
Richard P, Denjoy I, Fressart V, et al. Advising a cardiac disease gene positive yet phenotype negative or borderline abnormal athlete: is sporting disqualification really necessary? Br J Sports Med. 2012;46(Suppl 1):i59–68.
Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60(8):705–15.
Campuzano O, Alcalde M, Berne P, et al. Genetic testing of candidate genes in arrhythmogenic right ventricular cardiomyopathy/dysplasia. Eur J Med Genet. 2012;55(4):225–34.
• Elliott P, O’Mahony C, Syrris P, et al.: Prevalence of desmosomal protein gene mutations in patients with dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3(4):314–22.
Study demonstrating the prevalence of desmosomal mutations in DCM
Garcia-Pavia P, Syrris P, Salas C, et al. Desmosomal protein gene mutations in patients with idiopathic dilated cardiomyopathy undergoing cardiac transplantation: a clinicopathological study. Heart. 2011;97(21):1744–52.
• Meder B, Haas J, Keller A, et al.: Targeted next-generation sequencing for the molecular genetic diagnostics of cardiomyopathies. Circ Cardiovasc Genet. 2011;4(2):110–122.
Provides evidence on how NGS technologies can be used in diagnostics
Morales A, Painter T, Li R, et al. Rare variant mutations in pregnancy-associated or peripartum cardiomyopathy. Circulation. 2010;121(20):2176–82.
• Herman DS, Lam L, Taylor MR, et al.: Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366(7):619–28. Study reporting the presence of truncations of titin in DCM
Taylor M, Graw S, Sinagra G, et al. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation. 2011;124(8):876–85.
Galmiche L, Serre V, Beinat M, et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum Mutat. 2011;32(11):1225–31.
Götz A, Tyynismaa H, Euro L, et al. Exome sequencing identifies mitochondrial alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am J Hum Genet. 2011;88(5):635–42.
Norton N, Li D, Rieder MJ, Siegfried JD, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–82.
Villard E, Perret C, Gary F, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–76.
Theis JL, Sharpe KM, Matsumoto ME, et al. Homozygosity mapping and exome sequencing reveal GATAD1 mutation in autosomal recessive dilated cardiomyopathy. Circ Cardiovasc Genet. 2011;4(6):585–94.
• Cox MG, van der Zwaag PA, van der Werf C, et al.: Arrhythmogenic right ventricular dysplasia/cardiomyopathy: pathogenic desmosome mutations in index-patients predict outcome of family screening: Dutch arrhythmogenic right ventricular dysplasia/cardiomyopathy genotype-phenotype follow-up study. Circulation. 2011;123(23):2690–700.
This study demonstrates the utility of comprehensive documentation of genotypic and phenotypic data from a large series of probands to aid genotype-phenotype correlation
Pan S, Caleshu CA, Dunn KE, et al.: Cardiac structural and sarcomere genes associated with cardiomyopathy exhibit marked intolerance of genetic variation. Circ Cardiovasc Genet. 2012;5(6):602–10.
Hoage T, Ding Y, Xu X. Quantifying cardiac functions in embryonic and adult zebrafish. Methods Mol Biol. 2012;843:11–20.
Choi JC, Worman HJ.: Reactivation of autophagy ameliorates LMNA cardiomyopathy. Autophagy. 2012;9(1) [Epub ahead of print].
Purevjav E, Arimura T, Augustin S, et al. Molecular basis for clinical heterogeneity in inherited cardiomyopathies due to myopalladin mutations. Hum Mol Genet. 2012;21(9):2039–53.
Bagnall RD, Tsoutsman T, Shephard RE, et al. Global MicroRNA profiling of the mouse ventricles during development of severe hypertrophic cardiomyopathy and heart failure. PLoS One. 2012;7(9):e44744.
Manning EP, Guinto PJ, Tardiff JC, et al. Correlation of molecular and functional effects of mutations in cardiac troponin T linked to familial hypertrophic cardiomyopathy: an integrative in silico/in vitro approach. J Biol Chem. 2012;287(18):14515–23.
Jordan DM, Kiezun A, Baxter SM, et al. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am J Hum Genet. 2011;88(2):183–92.
•• Kapplinger JD, Landstrom AP, Salisbury BA, et al.: Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011;57(23):2317–27.
Comprehensive evaluation of genetic variation in healthy controls for the ARVC susceptibility genes
Page SP, Kounas S, Syrris P, et al. Cardiac myosin binding protein-C mutations in families with hypertrophic cardiomyopathy: disease expression in relation to age, gender, and long term outcome. Circ Cardiovasc Genet. 2012;5(2):156–66.
• Maron BJ, Maron MS, Semsarian C.: Double or compound sarcomere mutations in hypertrophic cardiomyopathy: a potential link to sudden death in the absence of conventional risk factors. Heart Rhythm. 2012;9(1):57–63.
Provides evidence for a potential gene-dosage effect in HCM
Xu T, Yang Z, Vatta M, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55(6):587–97.
Nakajima T, Kaneko Y, Irie T, et al. Compound and digenic heterozygosity in desmosome genes as a cause of arrhythmogenic right ventricular cardiomyopathy in Japanese patients. Circ J. 2012;76(3):737–43.
Quarta G, Muir A, Pantazis A, et al. Familial evaluation in arrhythmogenic right ventricular cardiomyopathy: impact of genetics and revised task force criteria. Circulation. 2011;123(23):2701–9.
van Rijsingen IA, Arbustini E, Elliott PM, et al. Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers a European cohort study. J Am Coll Cardiol. 2012;59(5):493–500.
• Quarta G, Syrris P, Ashworth M, et al.: Mutations in the Lamin A/C gene mimic arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2012;33(9):1128–36.
Provides evidence on lamin A/C in ARVC
van der Zwaag PA, van Rijsingen IA, Asimaki A, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14(11):1199–207.
Zhang M, Tavora F, Oliveira JB, et al. PKP2 mutations in sudden death from arrhythmogenic right ventricular cardiomyopathy (ARVC) and sudden unexpected death with negative autopsy (SUDNA). Circ J. 2012;76(1):189–94.
Maron BJ, Roberts WC, Arad M, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301(12):1253–9.
Ding Y, Sun X, Huang W, et al. Haploinsufficiency of target of rapamycin attenuates cardiomyopathies in adult zebrafish. Circ Res. 2011;109(6):658–69.
Marin TM, Keith K, Davies B, et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121(3):1026–43.
Kushwaha S, Xu X. Target of rapamycin (TOR)-based therapy for cardiomyopathy: evidence from zebrafish and human studies. Trends Cardiovasc Med. 2012;22(2):39–43.
Disclosure
The authors reported no potential conflicts of interest relevant to this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wijeyeratne, Y.D., Behr, E.R. Recent Developments in the Genetics of Cardiomyopathies. Curr Genet Med Rep 1, 21–29 (2013). https://doi.org/10.1007/s40142-012-0002-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40142-012-0002-2