Abstract
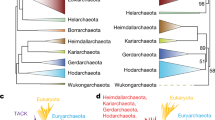
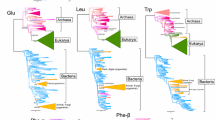
The origin of life is a long-standing mystery puzzling many people. This mystery possesses not only philosophical but also important biological significance. To unveil this mystery, the searches for the root of life, or the last universal common ancestor (LUCA), based on comparative-genomic analysis have been intensively performed on rRNAs, tRNAs and proteins sequences. The current search pointed to a Methanopyrus-proximal LUCA, which opens up the reconstruction of Lucan biology and helps to delineate the evolutionary pathways.
Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acid Res 25:3389–3402
Barns SM, Delwiche CF, Palmer JD, Pace NR (1996) Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA 93:9188–9193
Brinkmann H, Philippe H (1999) Archaea sister group of bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Bio Evol 16:817–825
Brown JR, Doolittle WF (1995) Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci USA 92:2441–2445
Burggraf S, Stetter KO, Rouviere P, Woese CR (1991) Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst Appl Microbiol 14:346–351
De Rijk P, Van de Peer Y, Van den Broeck I, De Wachter R (1995) Evolution according to large ribosomal subunit RNA. J Mol Evol 41:366–375. doi:10.1007/BF01215184
Delong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
Di Giulio M (1995) The phylogeny of tRNAs seems to confirm the predictions of the coevolution theory of the origin of the genetic code. Orig Life Evol Bisph 25:549–564. doi:10.1007/BF01582024
Di Giulio M (2000) The RNA world, the genetic code and tRNA molecule. Trends Genet 16:17–19
Di Giulio M (2007) The tree of life might be rooted in the branch leading to Nanoarchaeota. Gene 401:108–113. doi:10.1016/j.gene.2007.07.004
Doolittle WF (1999) Phylogenetic classification and the universal tree. Science 284:2124–2128. doi:10.1126/science.284.5423.2124
Fox GE, Magrum LJ, Balch WE, Wolfe RS, Woese CR (1977a) Classification of methanogenic bacterial by 16S ribosomal RNA characterization. Proc Natl Acad Sci USA 74:4537–4541
Fox GE, Peckman KJ, Woese CR (1977b) Comparative cataloging of 16S ribosomal ribonucleic acid; molecular approach to prokaryotic systematics. Int J Syst Bacteriol 27:44–57. doi:10.1099/00207713-27-1-44
Gogarten JP, Olendzenski L (1999) Orthologs, paralogs and genome comparisons. Cur Opin Genet Develop 9:630–636. doi:10.1016/S0959-437X(99)00029-5
Gogarten JP, Kibak H, Dittrich H, Taiz L, Bowman EJ, Manolson EJ, Poole RJ, Date T et al (1989) Evolution of vacuolar H+-ATPase: implications for the origin of eukarya. Proc Natl Acad Sci USA 86:6661–6665
Graur D, Li WH (2000) Molecular phylogenetics, chapter 5 and gene duplication, exon shuffling and concerted evolution. Chapter 6. In: Fundamentals of molecular evolution, 2nd edn. Sinauer, Sunderland
Gribaldo S, Philippe H (2002) Ancient phylogenetic relationships. Theor Popul Biol 61:391–408. doi:10.1006/tpbi.2002.1593
Hafenbradl D, Keller M, Thiericke R, Stetter KO (1993) A novel unsaturated archaeal ether core lipid from the hyperthermophile Methanopyrus kandleri. Syst Appl Microbiol 16:165–169
Hohn MJ, Hedlund BP, Huber H (2002) Detection of 16S rDNA sequences representing the novel phylum “Nanoarchaeota”: indication for a wide distribution in high temperature biotopes. Syst Appl Microbiol 25:551–554
Huber R, Kurr M, Jannasch HW, Stetter KO (1989) A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110°C. Nature 342:833–834. doi:10.1038/342833a0
Iwabe N, Kuma KI, Hasegawa M, Osawa S, Miyata T (1989) Evolutionary relationship of archaebacteria, eubacteria, and eukarya inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA 86:9355–9359
Kollman JM, Doolittle RF (2000) Determining the relative rates of change for prokaryotic and eukaryotic proteins with anciently duplicated paralogs. J Mol Evol 51:173–181. doi:10.1007/s002390010078
Lake JA, Servin JA, Herbold CW, Skophammer RG (2008) Evidence for a new root of the tree of life. Syst Biol 57:835–843. doi:10.1080/10635150802555933
Nesbo CL, Boucher Y, Doolittle WF (2001) Defining the core of non-transferable prokaryotic genes: the euryarchaeal core. J Mol Evol 53:340–350. doi:10.1007/s002390010224
Philippe H, Forterre P (1999) The rooting of universal tree of life is not reliable. J Mol Evol 49:509–523. doi:10.1007/PL00006573
Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR 3rd, Ibba M, Soll D (2005) RNA-dependent cysteine biosynthesis in Archaea. Science 307:1969–1972. doi:10.1126/science.1108329
Schwartz RM, Dayhoff MQ (1978) Origins of prokaryotes, eukaryotes, mitochondria and chloroplasts. Science 199:395–403. doi:10.1126/science.202030
Somerville CC, Knight IT, Straube WL, Colwell RR (1989) Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol 55:548–554
Takai K, Gamo T, Tsunogai U, Nakayama N, Hirayama H, Nealson KH, Horikoshi K (2004a) Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269–282. doi:10.1007/s00792-004-0386-3
Takai K, Oida H, Suzuki Y, Hirayama H, Nakagawa S, Nunoura T, Inagaki F, Nealson KH, Horikoshi K (2004b) Spatial distribution of marine Crenarchaeota Group I in the vicinity of deep-sea hydrothermal systems. Appl Environ Microbiol 70:2404–2413. doi:10.1128/AEM.70.4.2404-2413.2004
Van der Auwera G, Hofmann CJ, De Rijk P, De Wachter R (1998) The origin of red algae and cryptomonad nucleomorphs: a comparative phylogeny based on small and large subunit rRNA sequences of Palmaria palmata, Gracilaria verrucosa, and the Guillardia theta nucleomorph. Mol Phylogenet Evol 10:333–342. doi:10.1006/mpev.1998.0544
Woese CR (1987) Bacteria evolution. Microbiol Rev 51:221–271
Woese CR (1998) The universal ancestor. Proc Natl Acad Sci USA 95:6854–6859
Woese CR, Fox GE (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA 74:8392–8396
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc Natl Acad Sci USA 87:4576–4578
Wolf YI, Rogozin IB, Grishin NV, Koonin EV (2002) Genome trees and the tree of life. Trends Genet 18:472–479. doi:10.1016/S0168-9525(02)02744-0
Wong JT (1975) A coevolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Wong JT (1976) The evolution of a universal genetic code. Proc Natl Acad Sci USA 73:2336–2340
Wong JT (2005) Coevolution theory of the genetic code at age thirty. BioEssays 27:416–425. doi:10.1002/bies.20208
Wong JT (2009) Root of life. In: Wong JT, Lazcano A (eds) Prebiotic evolutuion and astrobiology. Landes Bioscience, Austin, pp 120–143
Wong JT, Xue H (2002) Self-perfecting evolution of heteropolymer building blocks and sequences as the basis of life. Chapter III 10. In: Fundamentals of life. Elsevier, Paris, pp 473–494
Wong JT, Chen J, Mat WK, Ng SK, Xue H (2007) Polyphasic evidence delineating the root of life and roots of biological domains. Gene 403:39–52. doi:10.1016/j.gene.2007.07.032
Xue H, Tong KL, Mark C, Grosjean H, Wong JT (2003) Transfer RNA paralogs: evidence for genetic code-amino acid biosynthesis coevolution and an archaeal root of life. Gene 310:59–66. doi:10.1016/s0378-1119(03)00552-3
Xue H, Ng SK, Tong KL, Wong JT (2005) Congruence of evidence for a Methanopyrus-proximal root of life based on transfer RNA and AARS genes. Gene 360:120–130. doi:10.1016/j.gene.2005.06.027
Yu Z (2010) Optimization of PCR amplification for sensitive capture of Methanopyrus isoleucyl-tRNA synthetase gene in environmental samples. Ann Microbiol 60:757–762. doi:10.1007/s13213-010-0097-1
Yu Z, Takai K, Slesarev A, Xue H, Wong JT (2009) Search for primitive Methanopyrus based on genetic distance between Val- and Ile-tRNA synthetases. J Mol Evol 69:386–394. doi:10.1007/s00239-009-9297-3
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yu, Z., Xu, S. Search for a Methanopyrus-proximal last universal common ancestor based on comparative-genomic analysis. Ann Microbiol 61, 397–401 (2011). https://doi.org/10.1007/s13213-010-0154-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-010-0154-9