Abstract
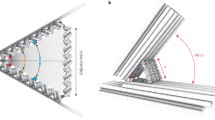
We previously demonstrated that polypod-like structured DNA, or polypodna, constructed with three or more oligodeoxynucleotides (ODNs), is efficiently taken up by immune cells such as dendritic cells and macrophages, depending on its structural complexity. The ODNs comprising the polypodna should bend to form the polypod-like structure, and may do so by adopting either a bendtype conformation or a cross-type conformation. Here, we tried to elucidate the orientation and bending of ODNs in polypodnas using atomic force microscopy (AFM). We designed two types of pentapodnas (i.e., a polypodna with five pods) using 60- to 88-base ODNs, which were then immobilized on DNA origami frames. AFM imaging showed that the ODNs in the pentapodna adopted bend-type conformations. Tetrapodna and hexapodna also adopted bend-type conformations when they were immobilized on frames under unconstrained conditions. These findings provide useful information toward the coherent design of, and the structure–activity relationships for, a variety of DNA nanostructures.

Similar content being viewed by others
References
Um, S. H.; Lee, J. B.; Park, N.; Kwon, S. Y.; Umbach, C. C.; Luo, D. Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006, 5, 797–801.
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302.
He, Y.; Ye, T.; Su, M.; Zhang, C.; Ribbe, A. E.; Jiang, W.; Mao, C. D. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 2008, 452, 198–201.
Smith, D; Schüller, V; Engst, C; Rädler, J; Liedl, T. Nucleic acid nanostructures for biomedical applications. Nanomedicine 2013, 8, 105–121.
Um, S. H.; Lee, J. B.; Kwon, S. Y.; Li, Y. G.; Luo, D. Dendrimer-like DNA-based fluorescence nanobarcodes. Nat. Protoc. 2006, 1, 995–1000.
Ke, Y. G.; Ong, L. L.; Shih, W. M.; Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 2012, 338, 1177–1183.
Rattanakiat, S.; Nishikawa, M.; Funabashi, H.; Luo, D.; Takakura, Y. The assembly of a short linear natural cytosinephosphate-guanine DNA into dendritic structures and its effect on immunostimulatory activity. Biomaterials 2009, 30, 5701–5706.
Mohri, K.; Nishikawa, M.; Takahashi, N.; Shiomi, T.; Matsuoka, N.; Ogawa, K.; Endo, M.; Hidaka, K.; Sugiyama, H.; Takahashi, Y. et al. Design and development of nanosized DNA assemblies in polypod-like structures as efficient vehicles for immunostimulatory CpG motifs to immune cells. ACS Nano 2012, 6, 5931–5940.
Wemmer, D. E.; Wand, A. J.; Seeman, N. C.; Kallenbach, N. R. NMR analysis of DNA junctions: Imino proton NMR studies of individual arms and intact junction. Biochemistry 1985, 24, 5745–5749.
Ortiz-Lombardía, M.; González, A.; Eritja, R.; Aymamí, J.; Azorín, F.; Coll, M. Crystal structure of a DNA Holliday junction. Nat. Struct. Biol. 1999, 6, 913–917.
Lilley, D. M. Structures of helical junctions in nucleic acids. Q Rev. Biophys. 2000, 33, 109–159.
Li, Y. G.; Tseng, Y. D.; Kwon, S. Y.; d’Espaux, L.; Bunch, J. S.; McEuen, P. L.; Luo, D. Controlled assembly of dendrimer-like DNA. Nat. Mater. 2004, 3, 38–42.
Mizuno, R.; Haruta, H.; Morii, T.; Okada, T.; Asakawa, T.; Hayashi, K. Synthesis and AFM visualization of DNA nanostructures. Thin Solid Films 2004, 459–465.
Lyubchenko, Y. L.; Shlyakhtenko, L. S.; Ando, T. Imaging of nucleic acids with atomic force microscopy. Methods 2011, 54, 274–283.
Nishikawa, M.; Mizuno, Y.; Mohri, K.; Matsuoka, N.; Rattanakiat, S.; Takahashi, Y.; Funabashi, H.; Luo, D.; Takakura, Y. Biodegradable CpG DNA hydrogels for sustained delivery of doxorubicin and immunostimulatory signals in tumor-bearing mice. Biomaterials 2011, 32, 488–494.
Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Regulation of DNA methylation using different tensions of double strands constructed in a defined DNA nanostructure. J. Am. Chem. Soc. 2010, 132, 1592–1597.
Suzuki, Y.; Endo, M.; Katsuda, Y.; Ou, K.; Hidaka, K.; Sugiyama, H. DNA origami based visualization system for studying site-specific recombination events. J. Am. Chem. Soc. 2014, 136, 211–218.
Murchie, A. I. H.; Clegg, R. M.; von Krtzing, E.; Duckett, D. R.; Diekmann, S.; Lilley, D. M. J. Fluorescence energy transfer shows that the four-way DNA junction is a righthanded cross of antiparallel molecules. Nature 1989, 341, 763–766.
Eichman, B. F.; Vargason, J. M.; Mooers, B. H. M.; Ho, P. S. The Holliday junction in an inverted repeat DNA sequence: Sequence effects on the structure of four-way junctions. Proc. Natl. Acad. Sci. USA 2000, 97, 3971–3976.
Sha, R.; Liu, F.; Seeman, N. C. Atomic force microscopic measurement of the interdomain angle in symmetric Holliday junctions. Biochemistry 2002, 41, 5950–5955.
Peters 3rd, J. P.; Maher, L. J. DNA curvature and flexibility in vitro and in vivo. Q Rev. Biophys. 2010, 43, 23–63.
Vafabakhsh, R.; Ha, T. Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science 2012, 337, 1097–1101.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Shiomi, T., Tan, M., Takahashi, N. et al. Atomic force microscopy analysis of orientation and bending of oligodeoxynucleotides in polypod-like structured DNA. Nano Res. 8, 3764–3771 (2015). https://doi.org/10.1007/s12274-015-0875-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12274-015-0875-y