Abstract
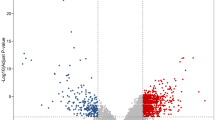
Colorectal cancer (CRC) is the most common malignant tumor of digestive system. The aim of this study was to identify gene signatures during CRC and uncover their potential mechanisms. The gene expression profiles of GSE21815 were downloaded from GEO database. The GSE21815 dataset contained 141 samples, including 132 CRC and 9 normal colon epitheliums. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analyses were performed, and protein–protein interaction (PPI) network of the differentially expressed genes (DEGs) was constructed by Cytoscape software. In total, 3500 DEGs were identified in CRC, including 1370 up-regulated genes and 2130 down-regulated genes. GO analysis results showed that up-regulated DEGs were significantly enriched in biological processes (BP), including cell cycle, cell division, and cell proliferation; the down-regulated DEGs were significantly enriched in biological processes, including immune response, intracellular signaling cascade and defense response. KEGG pathway analysis showed the up-regulated DEGs were enriched in cell cycle and DNA replication, while the down-regulated DEGs were enriched in drug metabolism, metabolism of xenobiotics by cytochrome P450, and retinol metabolism pathways. The top 10 hub genes, GNG2, AGT, SAA1, ADCY5, LPAR1, NMU, IL8, CXCL12, GNAI1, and CCR2 were identified from the PPI network, and sub-networks revealed these genes were involved in significant pathways, including G protein-coupled receptors signaling pathway, gastrin-CREB signaling pathway via PKC and MAPK, and extracellular matrix organization. In conclusion, the present study indicated that the identified DEGs and hub genes promote our understanding of the molecular mechanisms underlying the development of CRC, and might be used as molecular targets and diagnostic biomarkers for the treatment of CRC.


Similar content being viewed by others
References
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86.
Aran V, Victorino AP, Thuler LC, Ferreira CG. Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer. 2016;15(3):195–203.
Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–99.
Nannini M, Pantaleo MA, Maleddu A, et al. Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev. 2009;35:201–9.
Bustin SA, Dorudi S. Gene expression profiling for molecular staging and prognosis prediction in colorectal cancer. Expert Rev Mol Diagn. 2004;4:599–607.
Lascorz J, Hemminki K, Forsti A. Systematic enrichment analysis of gene expression profiling studies identifies consensus pathways implicated in colorectal cancer development. J Carcinog. 2011;10:7.
Iwaya T, Yokobori T, Nishida N, et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33:2391–7.
Gene Ontology C. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–6.
Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3.
Sakai E, Fukuyo M, Ohata K, et al. Genetic and epigenetic aberrations occurring in colorectal tumors associated with serrated pathway. Int J Cancer. 2016;138:1634–44.
Gautam V, Singh A, Singh S, Sarkar AK. An efficient LCM-based method for tissue specific expression analysis of genes and miRNAs. Sci Rep. 2016;6:21577.
Mehta P, Premkumar B, Morris R. Production of high quality brain-derived neurotrophic factor (BDNF) and tropomyosin receptor kinase B (TrkB) RNA from isolated populations of rat spinal cord motor neurons obtained by Laser Capture Microdissection (LCM). Neurosci Lett. 2016;627:132–8.
Perez R, Wu N, Klipfel AA, Beart RW Jr. A better cell cycle target for gene therapy of colorectal cancer: cyclin G. J Gastrointest Surg. 2003;7:884–9.
Tominaga O, Nita ME, Nagawa H, et al. Expressions of cell cycle regulators in human colorectal cancer cell lines. Jpn J Cancer Res. 1997;88:855–60.
Tsunoda T, Nakamura T, Ishimoto K, et al. Upregulated expression of angiogenesis genes and down regulation of cell cycle genes in human colorectal cancer tissue determined by cDNA macroarray. Anticancer Res. 2001;21:137–43.
Djamgoz MB, Coombes RC, Schwab A. Ion transport and cancer: from initiation to metastasis. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130092.
An N, Yang X, Zhang Y, et al. Cell cycle related genes up-regulated in human colorectal development predict the overall survival of late-stage colorectal cancer patients. Mol BioSyst. 2016;12:541–52.
Vicente CM, Ricci R, Nader HB, Toma L. Syndecan-2 is upregulated in colorectal cancer cells through interactions with extracellular matrix produced by stromal fibroblasts. BMC Cell Biol. 2013;14:25.
Chen D, Yu Z, Zhu Z, Lopez CD. The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells. Cancer Res. 2006;66:3485–94.
Shen L, Qu X, Ma Y, et al. Tumor suppressor NDRG2 tips the balance of oncogenic TGF-beta via EMT inhibition in colorectal cancer. Oncogenesis. 2014;3:e86.
Logan CV, Szabadkai G, Sharpe JA, et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat Genet. 2014;46:188–93.
Leung T, Chen H, Stauffer AM, et al. Zebrafish G protein gamma2 is required for VEGF signaling during angiogenesis. Blood. 2006;108:160–6.
Yajima I, Kumasaka MY, Yamanoshita O, et al. GNG2 inhibits invasion of human malignant melanoma cells with decreased FAK activity. Am J Cancer Res. 2014;4:182–8.
Citron M, Decker R, Chen S, et al. O6-methylguanine-DNA methyltransferase in human normal and tumor tissue from brain, lung, and ovary. Cancer Res. 1991;51:4131–4.
Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–71.
Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G: C to A: T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–92.
Meek RL, Urieli-Shoval S, Benditt EP. Expression of apolipoprotein serum amyloid A mRNA in human atherosclerotic lesions and cultured vascular cells: implications for serum amyloid A function. Proc Natl Acad Sci U S A. 1994;91:3186–90.
Yang M, Liu F, Higuchi K, et al. Serum amyloid A expression in the breast cancer tissue is associated with poor prognosis. Oncotarget. 2016. doi:10.18632/oncotarget.8561.
Ren Y, Wang H, Lu D, et al. Expression of serum amyloid A in uterine cervical cancer. Diagn Pathol. 2014;9:16.
Defer N, Best-Belpomme M, Hanoune J. Tissue specificity and physiological relevance of various isoforms of adenylyl cyclase. Am J Physiol Renal Physiol. 2000;279:F400–16.
Jarvandi S, Davidson NO, Schootman M. Increased risk of colorectal cancer in type 2 diabetes is independent of diet quality. PLoS ONE. 2013;8:e74616.
Wei JS, Johansson P, Chen L, et al. Massively parallel sequencing reveals an accumulation of de novo mutations and an activating mutation of LPAR1 in a patient with metastatic neuroblastoma. PLoS ONE. 2013;8:e77731.
Przygodzka P, Papiewska-Pajak I, Bogusz H, et al. Neuromedin U is upregulated by SNAIL at early stages of EMT in HT29 colon cancer cells. Biochim Biophys Acta. 2016. doi:10.1016/j.bbagen.2016.07.012.
Wang J, Wang Y, Wang S, et al. Bone marrow-derived mesenchymal stem cell-secreted IL-8 promotes the angiogenesis and growth of colorectal cancer. Oncotarget. 2015;6:42825–37.
Liu F, Yu C. IL-8 promote carcinogenesis of primary epithelial cells from familial adenomatous polyposis. Cell Biochem Biophys. 2014;70:1765–71.
Sotsios Y, Whittaker GC, Westwick J, Ward SG. The CXC chemokine stromal cell-derived factor activates a Gi-coupled phosphoinositide 3-kinase in T lymphocytes. J Immunol. 1999;163:5954–63.
Brand S, Dambacher J, Beigel F, et al. CXCR4 and CXCL12 are inversely expressed in colorectal cancer cells and modulate cancer cell migration, invasion and MMP-9 activation. Exp Cell Res. 2005;310:117–30.
Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697–710.
Zhao L, Lim SY, Gordon-Weeks AN, et al. Recruitment of a myeloid cell subset (CD11b/Gr1 mid) via CCL2/CCR2 promotes the development of colorectal cancer liver metastasis. Hepatology. 2013;57:829–39.
Chandrasekar B, Bysani S, Mummidi S. CXCL16 signals via Gi, phosphatidylinositol 3-kinase, Akt, I kappa B kinase, and nuclear factor-kappa B and induces cell-cell adhesion and aortic smooth muscle cell proliferation. J Biol Chem. 2004;279:3188–96.
Rieken S, Herroeder S, Sassmann A, et al. Lysophospholipids control integrin-dependent adhesion in splenic B cells through G(i) and G(12)/G(13) family G-proteins but not through G(q)/G(11). J Biol Chem. 2006;281:36985–92.
Foley JF, Singh SP, Cantu M, et al. Differentiation of human T cells alters their repertoire of G protein alpha-subunits. J Biol Chem. 2010;285:35537–50.
Damaghi M, Wojtkowiak JW, Gillies RJ. pH sensing and regulation in cancer. Front Physiol. 2013;4:370.
Siu YT, Jin DY. CREB–a real culprit in oncogenesis. FEBS J. 2007;274:3224–32.
Abramovitch R, Tavor E, Jacob-Hirsch J, et al. A pivotal role of cyclic AMP-responsive element binding protein in tumor progression. Cancer Res. 2004;64:1338–46.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Human participants and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Liang, B., Li, C. & Zhao, J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol 33, 111 (2016). https://doi.org/10.1007/s12032-016-0829-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-016-0829-6