Abstract
A complex interplay between pathogen and host determines the immune response during viral infection. A set of cytosolic sensors are expressed by immune cells to detect viral infection. NOD-like receptors (NLRs) comprise a large family of intracellular pattern recognition receptors. Members of the NLR family assemble into large multiprotein complexes, termed inflammasomes, which induce downstream immune responses to specific pathogens, environmental stimuli, and host cell damage. Inflammasomes are composed of cytoplasmic sensor molecules such as NLRP3 or absent in melanoma 2 (AIM2), the adaptor protein ASC (apoptosis-associated speck-like protein containing caspase recruitment domain), and the effector protein procaspase-1. The inflammasome operates as a platform for caspase-1 activation, resulting in caspase-1-dependent proteolytic maturation and secretion of interleukin (IL)-1β and IL-18. This, in turn, activates the expression of other immune genes and facilitates lymphocyte recruitment to the site of primary infection, thereby controlling invading pathogens. Moreover, inflammasomes counter viral replication and remove infected immune cells through an inflammatory cell death, program termed as pyroptosis. As a countermeasure, viral pathogens have evolved virulence factors to antagonise inflammasome pathways. In this review, we discuss the role of inflammasomes in sensing viral infection as well as the evasion strategies that viruses have developed to evade inflammasome-dependent immune responses. This information summarises our understanding of host defence mechanisms against viruses and highlights research areas that can provide new approaches to interfere in the pathogenesis of viral diseases.
Similar content being viewed by others
Introduction
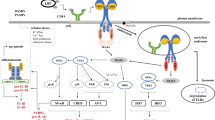
The innate immune system is the first line of defence differentiating ‘self’ from ‘non-self’. To detect and promptly respond to diverse groups of micro-organisms, the innate immune system appoints a range of germline-encoded pattern recognition receptors (PRRs) to detect pathogen-associated molecular patterns (PAMPs), which are conserved microbial components [1, 2]. PRRs are expressed by cells of the innate immune system such as macrophages, monocytes, dendritic cells, and neutrophils, as well as epithelial cells and cells of the adaptive immune system. A wide range of PRRs have been characterised, such as retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), Toll-like receptors (TLRs), nucleotide-binding oligomerisation domain (NOD)-like receptors (NLRs), and the absent in melanoma 2-like receptors (ALRs). Among these PRRs, TLRs detect PAMPs in the endosome and extracellular space, whereas NLRs, RLRs, and ALRs play a crucial role in sensing pathogens in the intracellular compartment (Fig. 1). The activation of PRRs results in the initiation of an innate immune response involving a cascade of signalling events that contribute to the differentiation and maturation of immune cells and the secretion of cytokines and chemokines, which also builds a base for the adaptive immune response [3].
Domain organisation and cellular localisation of pattern recognition receptors (PRRs). Toll-like receptors (TLRs), NOD-like receptors (NLRs), RIG-I-like receptors (RLRs), and AIM2-like receptors (ALRs) are displayed with their cellular localisation. TLRs are confined to the plasma membrane and endosomes. TLRs comprise leucine-rich repeat (LRR) domains and a Toll/interleukin-1 receptor (TIR) domain that facilitates downstream signalling. NLRs are present in the cytosol and are classified into five subfamilies based on their effector domain. Structurally, NLRs contain an N-terminal effector domain, a NACHT domain, and LRR domains. The effector domain can be an acidic activation domain (AD), baculovirus IAP repeat (BIR) domain, caspase recruitment domain (CARD), pyrin domain (PYD), or a domain without homology to other NLR subfamily members. RLRs are expressed in the cytoplasm and consist of CARD domains and an RNA helicase domain, except for LPG2, which lacks CARD domains. ALRs are localised in the cytoplasm and nucleus; they consist of a PYD domain and a certain number of HIN200 domains, which bind dsDNA
Cytosolic surveillance is performed by intracellular nucleic acid-sensing PRRs, including RNA-sensing RIG-like helicases (RIG-I and MDA5) and DNA-sensing PRRs (DAI and AIM2). The outcome of PAMP detection by PRRs is determined by the complex interplay between the invading microbe and responding cell. When the helicase domain of either RIG or MDA5 detects viral RNA in the cytoplasm, the caspase recruitment domain (CARD) is exposed to interact with the N-terminal of mitochondrial adaptor protein (MAVS) via its own CARD domain. This CARD–CARD interaction results in the dimerisation of MAVS in the mitochondria to form a protein complex called the MAVS signalosome, which enables the activation of NF-κB and the production of type I interferon (IFN). Once the innate immune system has been activated, it elicits the secretion of cytokines and chemokines, which subsequently induce the expression of adhesion molecules and costimulatory molecules to further activate the adaptive immune response [4–11].
In this context, the expression of proinflammatory cytokines such as interleukin (IL)-1β and IL-18 is induced by proinflammatory stimuli, including the activity of some PAMPs that can induce the synthesis of chemokines and adhesion molecules, resulting in the activation of a proinflammatory immune response [12]. Among these proinflammatory cytokines, IL-1β is involved in a range of biological processes such as apoptosis, pyroptosis, cell differentiation, proliferation, and regulation of the inflammatory immune response [13]. While the production of many proinflammatory cytokines is principally regulated at the transcriptional level, IL-1β and IL-18 require an additional proteolytic event that is regulated in two steps. The first regulatory step is stimulation via TLRs or RLRs, which induces the synthesis of IL-1β and IL-18 as inactive precursors. The second regulatory step involves post-translational processing; this is required for the secretion and bioactivity of these cytokines and is catalysed by inflammasome activation (Fig. 2) [14–16]. The inflammasome is a multiprotein complex that regulates the auto-cleavage process, which in turn activates caspase-1 and generates the p10/p20 tetramer. This further converts inactive IL-1β and IL-18 into their active forms [17]. The inflammasome plays an important role in the innate immune pathway and regulates at least two protective responses of the host: the secretion of proinflammatory cytokines (IL-1β and IL-18) and the induction of pyroptosis, which is a form of cell death [18, 19]. Although several studies have identified diverse inflammasomes with activities against a broad range of pathogens, certain inflammasomes, such as the NLRP3, AIM2, and RIG-I inflammasomes, have been found to be highly specific and important in mediating the host response to viral infection [11, 20]. During the past few years, several studies have demonstrated that certain viruses have the ability to activate inflammasomes, which in turn contributes to antiviral responses. In addition, studies in recent years have contributed towards an improved understanding of the molecular mechanisms underlying host immune evasion by viral pathogens, as well as the roles of NLRs and inflammasomes in mounting antiviral host responses. Here, we review the PRRs that constitute the multiprotein complexes called ‘Inflammasomes’ that regulate the maturation and secretion of IL-1β and IL-18. In this review, we will discuss the activation mechanism and roles of inflammasomes during viral infection along with the evolved mechanism through which viruses evade the antiviral innate immune response.
Molecular sensors associated with the antiviral innate immune response. Cells susceptible to viral infection have evolved a defence mechanism based on the recognition of pathogen-associated/danger-associated molecular patterns (PAMPs/DAMPs) of viral origin, including viral RNA and DNA. The recognition process is mediated by molecules distributed in several cellular compartments and culminates in the activation of NLR proteins and the inflammasome to initiate signalling and the production of proinflammatory cytokines, which leads to the amplification of the antiviral immune response. In the endosome, viral PAMPs are sensed by TLR-7 and TLR-3, and trigger a signal that activates the transcription factor NF-κB to promote the gene expression of cytokines including pro-IL-1β and pro-IL-18. In addition, RIG-1 recognises viral PAMPs via its helicase domain and further interacts with adapter proteins localised in mitochondria such as virus-induced signalling adapter, which contains CARD domains. This signalling culminates in the phosphorylation and activation of transcription factors such as IRF3 and IRF7, which in turn induce the transcription of type I INF and the production of IL-1β. Finally, after recognising viral PAMPs and DAMPs, NLRs (NLRP3) and AIM2 self-oligomerise and, via their PYD domains, recruit adapter molecules such as ASC that contain caspase recruitment domains. This mechanism triggers the activation of caspase-1, which processes pro-IL-1β and pro-IL-18 into their active forms
NLR family
The innate immune system is able to respond promptly against invading pathogens as a first line of defence. Sensing of microbial pathogens by the innate immune system relies on the specific host receptor detection of their molecular signatures, which comprise PAMPs and danger-associated molecular patterns (DAMPs). Host PRRs are germline-encoded and include a number of families of leucine-rich repeat (LRR)-bearing proteins in plants and animals. Recent studies have identified two protein families, the RIG-like helicases (RLHs) and the NLRs, which perform intracellular scrutiny for PAMPs and DAMPs, while the Toll-like receptors (TLRs) comprise key sensors for detection of extracellular or membrane-encased foreign organisms [21–23]. Furthermore, numerous NLR family proteins stimulate innate and adaptive immune responses [24]. NLRs initiate host defence after recognising microbial products or intracellular danger signals, through the activation of the NF-κB response and inflammatory caspases [25]. NLRs are characterised as central molecular platforms that establish signalling complexes such as inflammasomes and NOD signalosomes.
Structurally, the members of the Nod-like receptor (NLR) family are multi-domain proteins composed of a common central NOD domain, C-terminal LRRs, and N-terminal caspase recruitment ‘C’ (CARD) or pyrin ‘P’ (PYD) domains. LRRs play a crucial role in the processes of ligand sensing and autoregulation that initiate NLR signalling, whereas the CARD and PYD domains mediate homotypic protein–protein interactions for downstream signalling [26, 27]. Thus, the active multiprotein signalling platform (e.g. the inflammasome or nodosome) is formed by oligomerisation of the NACHT domains (which are present in all NLR family members) in an ATP-dependent manner, which further allows the attachment of adaptor molecules and effector proteins, eventually leading to an inflammatory response [27, 28]. Members of the NLR family have been further classified into 3 subfamilies based on the sequences of their NACHT domains: NODs (NOD1–2, NOD3/NLRC3, NOD4/NLRC5, NOD5/NLRX1, CIITA), NLRPs (NLRP1–14, also called NALPs [NLRP can refer to human genes that code for a series of N {NACHT}, L {LRR} and P {PYD} domain-containing proteins]), and IPAF (NLRC4 and NAIP) (NLRC can refer to human genes that code for a series of N (NACHT), L (LRR), and C (CARD) domain-containing proteins) (Fig. 1).
Inflammasome activation during viral infection
NLRP3 inflammasome
The NLRP3 inflammasome is the best-studied inflammasome and seems to be activated by many families of viruses, suggesting a common pathway for viral detection and response by the host cell [29]. The structural arrangement of NLRP3 (the order of its domains from N-terminus to C-terminus) is PYD–NOD–LRRs. Structurally, the NLRP3 inflammasome comprises the adaptor protein apoptosis-associated speck-like protein containing CARD (ASC) and procaspase-1. NLRP3 forms a homo-oligomer during activation via its NOD domain and further interacts with ASC through its PYD domain. ASC, in turn, interacts with procaspase-1 through its CARD domain, forming the complete NLRP3 inflammasome complex [30]. The activation of NLRP3 inflammasome leads to the activation of caspase-1 and the production of mature IL-1β and IL-18 [31]. For its full activation, the NLRP3 inflammasome requires two signals. The first signal (priming signal) can be produced by receptors of the TLR, NLR, or RIG-I-like receptor families, or by a cytokine receptor; this first signal leads to transcriptional activation of NLRP3, pro-caspase-1, and the genes encoding pro-IL-1β and pro-IL-18 [32]. The second signal for inflammasome activation occurs in response to infection, or various stress signals associated with host sterile cell/tissue damage [33]. Three key mechanisms are associated with NLRP3 inflammasome activation (Fig. 3). Firstly, NLRP3 activators such as alum, silica, nigericin, and asbestos stimulate the production of reactive oxygen species (ROS) by nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, which stimulates an efflux of potassium ions (K+) leading to the activation of NLRP3 inflammasome [34, 35]. The second mechanism is the ‘lysosomal rupture model’ in which NLRP3 activators (silica, asbestos, etc.) primarily form aggregates, which are phagocytosed and induce the release of cathepsin B by lysosome rupture. Released cathepsin B further activates NLRP3 [34]. Lastly, the ‘ion channel model’ posits that a high concentration of extracellular ATP can induce the formation of cell membrane pannexin-1 pores and K+ efflux through the P2X7 ATP-gated ion channel, assisting the influx of PAMPs and DAMPs, which play a crucial role in NLRP3 activation [36]. However, intracellular ion fluxes have been found to be the most crucial danger signal and amplify inflammasome recruitment. Several studies have demonstrated that in response to a broad range of stimuli, including ultraviolet light irradiation, membrane attack complex formation generates pores that allow the influx of calcium ions (Ca2+), which increases the cytosolic Ca2+ concentration [37–39]. Further, Ca2+ accumulation in the mitochondrial matrix leads to the loss of mitochondrial membrane potential, triggering NLRP3 activation and secretion [40]. Recently, the role of calcium in inflammasome activation has been shown in two different studies. In vitro, monocytes were shown to activate NLRP3 inflammasome when stimulated with extracellular calcium (CaCl2) in culture media. Further, the activation was found to be mediated by signalling through the calcium-sensing receptor (CaSR) and G protein-coupled calcium-sensing receptor (GPRC6A) via the phosphatidyl inositol/Ca2+ pathway. In addition, in the mouse model of carrageenan-induced footpad swelling, increased calcium concentration was found to amplify the inflammatory response and this effect was inhibited in Gprc6a −/− mice [41]. This work was further supported by the finding that extracellular Ca2+ induced increase in intracellular Ca2+ levels, triggering inflammasome activation [38]. It was observed that treatment of CaSR agonist in bone marrow-derived macrophages (BMDMs) triggers the activation of NLRP3 inflammasome, which indicates that the recognition of Ca2+ by murine CaSR activates NLRP3 inflammasome [38]. Additionally, it has been demonstrated that CaSR stimulates the NLRP3 inflammasome via phospholipase C (PLC), which catalyses inositol-1,4,5-trisphosphate production and thereby induces release of Ca2+ from endoplasmic reticulum stores. In this context, LPS-primed BMDMs treated with PLC activators showed elevated IL-1β secretion, in contrast to BMDMs treated with PLC inhibitor that blocked IL-1β secretion in the presence of either extracellular Ca2+ or ATP [38]. Subsequently, elevation of cytoplasmic Ca2+ levels triggered the assembly of NLRP3 inflammasome components [38].
Activation mechanisms of the inflammasome. (A) Mechanisms of virus-mediated NLRP3 activation during virus infection through viral genomic RNA (vRNA), which can be single-stranded RNA (ssRNA) or double-stranded RNA (dsRNA) depending on the type of virus, and by viroporin proteins, which induce ion efflux from intracellular storage into the cytosol and activate NLRP3 inflammasome. (B) High concentrations of extracellular ATP induce the efflux of potassium ions through the P2X7 ATP-gated ion channel, which allows more PAMPs and DAMPs to enter cells via pannexin-1 pores. (C) PAMPs and DAMPs can be phagocytosed resulting in the disruption of lysosomes and the consequent release of lysosomal components, especially cathepsin B. This then activates the NLRP3 inflammasome. (D) PAMPs and DAMPs induce the production of reactive oxygen species (ROS), which may activate thioredoxin-interacting protein (TXNIP) to trigger NLRP3 inflammasome activation. Finally, all inflammasome types (NLRP3, AIM2, and RIG-1) induce caspase-1 activation and interleukin (IL)-1β and IL-18 production via the adaptor molecule ASC, which is required to bridge the interaction between pattern recognition receptors (via its PYD domain) and caspases (via its CARD domain)
In addition, K+ efflux has also been shown to trigger inflammasome activation in response to several stimuli including mitochondrial and lysosomal damage, ROS, and extracellular Ca2+. These findings indicate that inflammasomes are stringently regulated by intracellular ion concentrations and ion imbalances serve as the central trigger for their activation [42]. A growing body of evidence regarding the mechanisms of inflammasome activation suggests that in several viral infections, viral products such as viroporins may alter cell membrane permeability and change the ionic milieu of cell membranes. Such changes in the properties of the cell membrane can play a significant role in NLRP3 inflammasome activation [43, 44] (Fig. 3).
RNA viruses
Cumulative work has demonstrated that many RNA viruses are able to activate ‘the NLRP3 inflammasome’. The first evidence came from an in vitro study on flock house virus, rotavirus, Sendai virus, and influenza A virus, which demonstrated that viral dsRNA activates caspase-1 in murine macrophages [45]. Later, other RNA viruses, such as encephalomyocarditis virus (ECMV) and vesicular stomatitis virus (VSV), were studied in murine dendritic cells (DCs) and macrophages, and the results indicated that IL-1β secretion was NLRP3-dependent [46]. Further, no production of IL-1β was observed when the cells were incubated with ultraviolet light-treated (i.e. inactivated) virions, suggesting that inflammation activation requires viral replication. Moreover, the survival rates of caspase-1-deficient and wild-type mice did not differ upon infection with EMCV and VSV, indicating the minor role of IL-1β in the host defence against these viruses [46]. Recently, it was shown that EMCV viroporin 2B, but not viral RNA, can activate the NLRP3 inflammasome [47]. Upon the expression of EMCV viroporin 2B, NLRP3 was redistributed to the perinuclear space, where it colocalises with viroporin 2B. In addition, viroporin 2B increases the Ca2+ concentration in the cytoplasm, promoting IL-1β secretion [47]. Measles virus infection was found to activate the NLRP3 inflammasome in THP-1 cells, resulting in the secretion of IL-1β [48]. Human rhinovirus (HRV) and its viroporin 2B protein activate the NLRP3 and NLR family CARD-containing 5 (NLRC5) inflammasome [49]. When bronchial cells were infected or transfected with HRV and its viroporin 2B protein, respectively, NLRP3 was found to interact with NLRC5 and ASC and colocalised with viroporin 2B and NLRC5 in the Golgi apparatus. Viroporin 2B of HRV leads to the activation of NLRP3 inflammasome by increasing the cytosolic Ca2+ concentration as treatment with either BAPTA-AM (a Ca2+ chelator) or verapamil (a Ca2+ channel inhibitor) can block HRV-induced IL-1β release and caspase-1 activation [50]. Previous studies in Huh7.5 hepatoma cells or THP-1 macrophages demonstrated that hepatitis C virus (HCV) induces IL-1β production and NLRP3 inflammasome activation in a ROS- and K+ channel-dependent manner [50, 51]. More specifically, it has been shown that HCV genomic RNA and viroporin P7 protein activate inflammasome activation, and its activation was found to be ASC- and NLRP3-dependent [52].
Among all RNA viruses, influenza virus is the best studied in regard to its capabilities to interact with and activate the NLRP3 inflammasome [53, 54]. Studies investigating the role of the NLRP3 inflammasome in the development of the adaptive immune response have yielded controversial results; the development of adaptive immunity was found to be mainly dependent on inflammasome activation, but was NLRP3-independent [55, 56]. Recently, studies on the mechanism underlying NLRP3 activation during influenza virus infection have suggested that the priming signal for activation of the NLRP3 inflammasome is provided following the recognition of viral RNA by TLR7, with subsequent IL-1β production [57]. Influenza virus provides the second signal for NLRP3 inflammasome activation by inserting its ion channel protein M2 into the trans-Golgi network. This disturbs the ionic milieu, leading to K+ efflux and ROS production, and ultimately resulting in the activation of the NLRP3 inflammasome [57]. In addition, secretion of IL-1β and IL-18 by blood bone marrow-derived macrophages (BMMs) was observed when the cells were infected with wild-type influenza virus, but not when they were infected with a mutated influenza virus expressing non-functional M2 protein. Furthermore, LPS-primed BMMs and blood bone marrow-derived DCs were found to activate the NLRP3 inflammasome when they were transduced with a recombinant lentivirus expressing the M2 protein. Therefore, it has been suggested that the ion channel activity of M2 protein plays a crucial role in the activation of the NLRP3 inflammasome [57]. Other RNA viruses such as respiratory syncytial virus (RSV) have been found to activate the NLRP3 inflammasome, resulting in the activation of caspase-1 and release of IL-1β. In this case, NLRP3 inflammasome activation was ROS- and K+ efflux-dependent [58].
In addition to ion channel proteins, RNA, and viral pore-forming toxins, pathogenic influenza A virus protein PB1-F2 has been demonstrated to activate caspase-1 and NLRP3-specific inflammasome. Furthermore, infection in mice with PB1-F2-deficient influenza A virus resulted in decreased IL-1β secretion in contrast to that with wild-type virus. In addition, mice have shown enhanced IL-1β secretion when exposed to PB1-F2 peptide derived from pathogenic Influenza A virus compared to mice exposed to peptide derived from seasonal Influenza A virus [59]. Additionally, one study demonstrated that viroporin SH from RSV provides the second signal for activation of the NLRP3 inflammasome; a mutant RSV lacking the viroporin SH gene was unable to induce caspase-1 maturation and IL-1β release, in contrast to the wild-type RSV. Further, treatment with 5-(N,N-hexamethylene) amiloride, which is an inhibitor of the HCV p7 channel, together with a Na+/H+ ion channel inhibitor was shown to inhibit the secretion of IL-1β [60]. These findings suggest that NLRP3 can be activated by virus-encoded viroporins due to their ability to disrupt the cellular ion balance. In the context of human immunodeficiency virus (HIV) infection, NLRP3 inflammasome activation is observed during the early onset of the HIV disease and HIV infection has been shown to activate the NLRP3 inflammasome and IL-1β secretion in DCs from healthy individuals, but not in those from HIV-positive individuals [61]. In addition to the aforementioned viruses, several other RNA viruses have been reported to activate the NLRP3 inflammasome.
A recent study on mice infected with West Nile virus (WNV) demonstrated that the infection is able to induce IL-1β secretion, caspase-1 maturation, and ASC expression. Furthermore, high mortality rates were observed in WNV-infected knockdown mice lacking functional Il-1r, Nlrp3, Casp-1, and Asc genes, while lower mortality rates were observed in WNV-infected wild-type mice [62, 63]. Additionally, mice infected with other members of the family Flaviviridae, such as Japanese encephalitis virus, show IL-1β and IL-18 secretion and caspase-1 maturation in an NLRP3-dependent, K+- and ROS-mediated manner. Furthermore, NLRP3 depletion results in reductions in caspase-1 activity and of IL-1β and IL-18 secretion [64]. Dengue virus (DENV) also induces the NLRP3 inflammasome in infected platelets in vitro and in those obtained from DENV-infected patients. This activation was found to be ROS- and K+ channel-mediated. IL-1β secretion from platelets plays a crucial role in increased human microvascular endothelial cell permeability in vitro and vascular permeability in clinical cases [65]. Further investigation regarding the mechanism of inflammasome activation by DENV demonstrated that C-type lectin domain family 5 members A (CLEC5A) interacts directly with DENV, triggering NLRP3 inflammasome activation [66]. In addition, Rift Valley fever virus (RVFV) also induces the formation of an inflammasome complex containing NLRP3 and MAVS in which MAVS localises with NLRP3 during the RVFV infection, and MAVS knockout mice showed a reduced production of IL-1β in bone marrow-derived DCs infected with RVFV [67]. Recently, a study on human chikungunya virus (CHIKV) infection showed a high expression of NLRP3 and caspase-1 activation that led to the secretion of IL-1β [68].
DNA viruses
NLRP3 senses both non-enveloped and enveloped DNA viruses [69, 70]. Studies on non-enveloped viruses such as adenovirus have shown conflicting results regarding the role of the inflammasome. Initially, Nlrp3-dependent inflammasome activation was demonstrated in murine macrophages transduced with adenovirus DNA or infected with adenovirus [69]. Further, NLRP3-independent sensing of adenovirus was observed, in which IL-1α and b3 integrins play a crucial role in triggering an early proinflammatory immune response [71]. Another study on human macrophages showed that NLRP3 activation occurs in a cathepsin B- and ROS-dependent manner during adenovirus infection [72, 73]. Infection in BMDMs with modified vaccinia virus Ankara (MVA) has demonstrated that both activation of inflammasome and transcription of IL-1β and NLRP3 are dependent on the crosstalk between TLR2-MyD88 and the NLRP3 inflammasome. [70]. Myxoma virus, a DNA virus of the genus Leporipoxvirus, has been shown to activate the NLRP3 inflammasome via the induction of ROS and cathepsin B. Additionally, varicella zoster virus activates NLRP3 inflammasome in macrophages, primary lung fibroblasts, and melanoma cells, resulting in the secretion of IL-1β and maturation of caspase-1 [74]. Thus, cumulative evidence strongly suggests that the NLRP3 inflammasome detects RNA and DNA viruses. However, the exact mechanisms of its activation and its relevance to viral infection and host antiviral innate immunity have not been elucidated.
The above evidence cumulatively indicates that sensing the infection of either RNA or DNA virus activates the NLRP3 inflammasome. Further, the precise mechanism and in vivo significance of NLRP3 inflammasome activation in response to viral infections remains elusive and requires more precise investigation.
AIM2 inflammasome
AIM2 belongs to the hematopoietic IFN-inducible nuclear proteins with a 200 amino acid repeat family (HIN200 family, also known as the IFI200 family), and contains a PYD domain and a C-terminal HIN200 domain. AIM2 activates downstream signalling by sensing cytoplasmic bacterial, viral, or even host double-stranded DNA (dsDNA) [16, 75, 76]. AIM2 does not contain a NOD domain, but it contains a C-terminal HIN200 domain, through which it interacts with dsDNA. Through its PYD domain, it interacts with ASC and thereby recruits caspase-1 to form the AIM2 inflammasome [16]. It has been shown that vaccinia virus (VACV) dsDNA induces both AIM2-dependent and NLRP3-independent maturation of caspase-1 [16]. In this regard, a study on cultured cells found that VACV and mouse cytomegalovirus (MCVM) could induce the maturation of caspase-1 in an AIM2-dependent manner. Furthermore, Aim2 −/− mice infected with MCVM demonstrated a blockade of IL-18 secretion, low IFNγ-producing natural killer cells, and an increase in MCVM viral titre in the blood as compared to wild-type mice [77]. Therefore, the available data strongly suggest that AIM2 induces the biosynthesis of proinflammatory cytokines to eliminate virus-infected cells through pyroptosis; thus, AIM2 might play a crucial role in clearing some viral infections [77]. Further, it has been proposed that AIM2 could be a sensor for viruses that produce DNA during their lifecycle, such as adenovirus and perhaps single-stranded RNA (ssRNA) retroviruses, which reverse-transcribe their RNA into dsDNA in the cytoplasm of infected cells. In contrast, herpes simplex virus 1 has been shown to activate the AIM2-independent inflammasome. This indicates that either AIM2 does not sense all dsDNA or certain viruses have evolved a specific mechanism to inhibit the recognition of their genomic DNA by AIM2 [77]. Interestingly, although earlier work suggested that AIM2 only recognises dsDNA, recent data have shown that CHIKV and WNV (both RNA viruses) induce the expression of AIM2 in primary dermal fibroblasts. Likewise, inhibition of AIM2 with siRNA has been shown to interfere with IL-1β production when primary dermal fibroblast cells are infected with CHIKV or WNV [78]. Furthermore, Zika virus (an ssRNA virus) is also able to stimulate AIM2 expression and subsequently the secretion of IL-1β in primary human skin fibroblasts [79]. More studies are required to understand the mechanism through which AIM2 recognises RNA viruses and induces inflammasome formation/activation and the subsequent maturation and secretion of caspase-1 and IL-1β, respectively. The findings discussed above reflect the significance of the AIM2 inflammasome in the induction of innate immune responses.
RIG-1
RIG-1 belongs to the RLR family and induces type 1 IFN production. The RIG-1 signalling process is initiated by the interaction of RIG-1 with MAVS. This interaction leads to the recruitment of tumour necrosis factor (TNF) receptor-associated factor 3 (TRAF-3) to activate the TRAF family member-associated nuclear factor-κB (NF-κB) activator-binding kinase (TBK)-1 complex. Further, it activates the TBK-1 complex to phosphorylate IFN response factor-3 and factor-7 (IRF-3 and IRF-7) to induce the production of type I IFN [80, 81]. Besides, the expression of proinflammatory cytokines is induced via RIG-1/MAVS interaction through the recruitment of TRAF-6 to activate NF-κB via the IκB kinase (IKK) complex [80, 81]. Structurally, RIG-1 contains two N-terminal CARD domains, a central RNA helicase domain and a C-terminal regulatory domain. The CARD domains interact with adaptor proteins to trigger downstream signalling [82]. The C-terminal regulatory domain binds with dsRNA resulting in conformational changes that lead to its oligomerisation and the subsequent interaction of RIG-1 with MAVS through a CARD–CARD interaction [80, 83]. The role of RIG-1 in the production of type 1 IFNs has been demonstrated in numerous viruses including Newcastle disease virus, VSV, Sendai virus, HCV, Japanese encephalitis virus, influenza A virus, rabies virus, measles virus, and RSV [84–91]. Further, a role of RIG-1 in triggering the immune response has been demonstrated for numerous viruses including DENV serotype 2, WNV, RSV, and HCV [86, 87, 92–94]. It has been demonstrated that rhabdovirus VSV infection in murine DCs induces RIG-1-dependent production of IL-1β and IL-18 via NF-κB, caspase-1, and caspase-3 activation; thus, RIG-1 is an inflammasome activator. Furthermore, DCs from RIG-1 knockdown mice are not able to secrete IL-1β [95]. However, the results of this study conflict with that of another report, which demonstrated that VSV is recognised by NLRP3 and not by RIG-1 [46]. These contrasting results need to be investigated further. Fascinatingly, these findings suggest the existence of dual roles for RIG-1 in the inflammasome and type 1 IFN pathways. A more recent study on primary human bronchial epithelial cells infected with influenza showed RIG-I-dependent priming of the NLRP3 inflammasome as well as direct RIG-I-mediated inflammasome activation [4]. Therefore, it can be assumed that RIG-I could activate its own inflammasome in response to some viruses, but its major functions in RIG-I-MAVS signalling and NLRP3 inflammasome activation are still the subject of debate, and further research is vitally needed.
NLRC5
NLRC5, an important regulator of MHC class I gene expression, has been extensively investigated in recent years in the context of virus infection. The role of NLRC5 in innate immunity is still not well understood, but it is known to participate in both the innate and adaptive immune responses [96–100]. In HeLa and THP-1 cells, NLRC5 has been demonstrated to positively regulate the IFN pathway, whereas in other models such as HEK 293 cells and RAW 264.7 cells, NLRC5 has been demonstrated to negatively regulate the IFN, NF-κB, and AP-1 pathways [96–98, 100]. In RAW 264.7 cells, it was demonstrated that the interaction of NLRC5 with IKKα results in the inhibition of its catalytic activity. NLRC5 binds RIG-I, and this interaction is critical for robust antiviral responses against influenza virus initiated by increased levels of TNF-α, IL-6, and IL-1β in the absence of NLRC5 [97]. In another study, NLRC5 was found to function as a trans-activator of MHC class I gene in HEK 293 cells, probably through the regulation of chromatin remodelling [99]. Further, a contrasting result was shown, wherein NLRC5 knockdown by RNAi was found to induce MHC class I expression [96]. The discrepancies between the works described above suggest that NLRC5-mediated host responses vary greatly among cell types. Recently, NRLC5- and NRLP3-dependent inflammasome activation was also shown to occur due to viroporin ion channel activity. In this report, rhinovirus ion channel protein 2B was shown to disturb intracellular Ca2+ homeostasis by inducing the release of Ca2+ from the Golgi body and endoplasmic reticulum, resulting in the activation of both the NLRP3 and NLRC5 inflammasomes. These results indicate that NLRP3 and NLRC5 act in a cooperative manner during inflammasome assembly by detecting intracellular Ca2+ fluxes and triggering IL-1β secretion [49]. Further reports have confirmed the synergy between NLRC5 and NLRP3 in macrophage cells, demonstrating that caspase-1 activity and the subsequent processing of pro-IL-1β and pro-IL-18 in response to NLRP3-specific agonists were inhibited in the absence of NLRC5. Further, self-association of NLRC5 has also been observed to induce inflammasome activation [101].
NLRX1
NLRX1 belongs to the family of NLR that contains the N-terminal domain with mitochondrion-targeting sequence, followed by NACHT and LRRs [102, 103]. NLRXI is unique among NLRs due to its localisation to mitochondria that probably makes the NLRX1 as a link between mitochondria function and innate immunity. NLRX1 interacts with CARD domain of MAVS through its NACHT domain on the cytosolic side of the mitochondrial outer membrane. This interaction leads to the obstruction of the RLH–MAVS interaction resulting in inhibition of MAVS-dependent antiviral responses, i.e. suppression of IFN-β and NF-κB. Further, it has been demonstrated that the C-terminal LRR domain of NLRX1 is required for MAVS-induced interferon. Additionally, Sendai virus infection has been shown to induce MAVS and RIG-1 interaction, which further mediates IRF3 dimer formation. This enhanced interaction between MAVS and RIG-1 and IRF3 dimer formation is attenuated in the presence of NLRX1, further supporting the negative regulatory role of NLRX1 in MAVS-mediated antiviral responses [102]. However, the interaction between NLRX1 and MAVS remains doubtful owing to the localisation of these two proteins in distinct mitochondrial subdomains that are separated by two membranes. Further, the overexpression of NLRX1 in mitochondria leads to ROS induction likely due to the interaction with UQCRC2, a matrix-facing component of the mitochondrial respiratory chain complex III that has a crucial role in ROS generation [104]. NLRX1 leads to ROS induction in synergy with proinflammatory stimuli such as Poly (I:C), Shigella, and TNF-α, which subsequently potentiates NF-κB and JNK pathways [103]. During viral infection, NLRX1 may have a role as the positive regulator of autophagy by interacting with the mitochondrial Tu translation elongation factor (TUFM) and mitochondrial immune signalling complex (MISC), which also includes ATG5, ATG12, and ATG16L1. TUFM further interacts with Atg5-Atg12 and Atg16L1, promotes autophagy, and shares the function of NLRX1 in inhibiting RLR-induced IFN-I, given that increased IFN-I and decreased autophagy has been demonstrated during stomatitis virus infection in the absence of NLRX1, which provides an advantage for host defence against vesicular stomatitis virus infection [105, 106]. Recently, NLRX1 has shown to defend the macrophage cells from apoptosis due to influenza virus by interacting with apoptotic protein PB1-F2 and promoting IFN signalling. Additionally, high viral titre and lower production of IFN is observed in NLRX1 knockout mice infected with influenza virus compared with those in the wild-type mice. This discrepancy of NLRX1 in positively regulating IFN in macrophage cells infected with influenza virus is owing to the ability of NLRX1 to interact strongly with viral protein PB1-F2 and prevent mitochondrial damage. Additionally, NLRX1-dependent production of type 1 IFN in macrophages is attenuated in the absence of protein PB1-F2, suggesting that NLRX1 is critical for macrophage immunity against influenza virus infection [107]. During infection by rhinovirus, NLRX1 has been shown to interact with viral RNA that recruits NLRX1 in the mitochondria. Additionally, ROS production and NADPH oxidase 1 induction during rhinovirus infection were found to be NLRX1-dependent [108]. Recently, NLRX1 has been shown to function as a negative regulator of the host immune response to HIV-1 and DNA viruses. Furthermore, NLRX1 has been demonstrated to inhibit STING signalling by blocking the interaction between STING and TANK-binding kinase 1 (TBK1), which is required for the induction of IFN-1 in response to DNA. Moreover, NLRX1−/− cells express a higher level of type 1 interferon during HIV-1 infection or treatment with cyclic GAMP or cyclic di-GMP, which triggers STING-dependent responses [109]. However, NLRX1 regulation is quite complex and seems to occur via cell type- and signal-dependent mechanisms; the above findings suggest the role of NLRX1 in the regulation of IFN during viral infection as well as the regulation and activation of inflammasomes. Investigation should be encouraged to understand the role of NLRX1 in the regulation of inflammasomes, since inhibiting NLRX1 activity might enhance the antiviral response.
Regulation of inflammasome activity and IL-1β secretion
Pyroptosis
Pyroptosis is a form of programmed cell death that depends on the activation of atleast one of the inflammatory caspases such as caspase-1 and caspase-11 in mice, and caspase-1, caspase-4, and caspase-5 in humans. The term pyroptosis comes from the Greek words ‘pyro’, which means fire (denoting the release of proinflammatory mediators) and ‘ptosis’, which means falling, a term commonly used to describe cell death [110]. Pyroptosis is initiated by caspase-1 following its activation by diverse inflammasomes, resulting in the lysis of infected cells. The activation of inflammatory caspases results in the formation of pores in the plasmatic membrane that measure approximately 1.1–2.4 μm in diameter, leading to an increase in cellular permeability [111]. Thus, the features of pyroptosis include rapid plasma membrane rupture and the release of intracellular proinflammatory contents such as DAMPs and cytokines, which induce a robust inflammatory response [110]. Destruction of the actin cytoskeleton has also been observed in pyroptotic cells, but the mechanism and importance of this event remain unclear [18, 112, 113]. Furthermore, during pyroptosis, DNA fragmentation and nuclear condensation occur [13, 18, 114]. Pyroptosis can efficiently remove infected cells and further stimulates the activation of the immune system via the release of pathogens from the dying cells. Furthermore, pyroptosis, which is associated with a highly inflammatory state induced by intracellular pathogens, has been reported to possibly contribute to an inflammatory syndrome in vivo [115]. In the non-canonical inflammasome pathway, it has been demonstrated that upon intracellular lipopolysaccharide (LPS) stimulation, caspase-11 cleaves the pannexin-1 channel resulting in the release of ATP, which in turn activates P2X7 to induce cytotoxicity and pyroptosis [116, 117]. Recent findings have suggested that the presence of cytosolic LPS dramatically decreases the threshold concentration of ATP necessary to drive the opening of the P2X7 channel, which allows the release of proinflammatory cytokines such as IL-1β. This further leads to P2X7 forming a receptor pore, allowing it to respond to the nanomolar concentration of released ATP. During the course of pyroptosis, ATP mostly works as an autocrine signal that targets the cells primed with hypersensitive P2X7 channels (such as those in which cytosolic LPS were detected) [116]. In addition, pannexin-1 was suggested to be necessary for mediating IL-1β release due to caspase-11 activation triggered by pannexin-1-mediated K+ efflux, which also drives assembly of the NLRP3 inflammasome [18]. Additionally, findings regarding the association between ASC and pyroptosis have suggested that the release of ASC specks from macrophages is pyroptosis-dependent [118, 119].
Negative regulation of inflammasome assembly
Two groups of proteins can act as negative modulators of inflammasome assembly and caspase-1-dependent production of IL-1β: PYD-only protein (POPs) and CARD-only proteins (COPs) [120]. POPs have been characterised as inflammasome regulators that interact and bind with PYD in ASC- and PYD-containing PRRs. Therefore, POPs inhibit the PYD–PYD interaction required for the recruitment and oligomerisation of ASC and consequently prevent inflammasome activation [120]. POP1 has been implicated in the regulation of inflammation by interacting with the PYD of ASC and suppressing NF-κB activity [113]. Moreover, findings have suggested that the suppression of NF-κB signalling by POP1 is mediated at the level of IKK via POP1 associating with IκBα and IKKβ, and inhibiting their kinase activities [121]. Thus, POP1 prevents inflammasome activation by obstructing ASC assembly, resulting in the inhibition of caspase-1 activation as well as IL-1β and IL-18 secretion, and weak nucleation of ASC polymerisation [118]. Another POP family member, called POP2, blocks TNF-α-mediated NF-κB activation [122]. POP2 acts distally in the NF-κB cascade by regulating the transactivation potential of the C-terminal transcriptional activation domain 1 of p65/RelA [123]. Although POP2 interacts with ASC and inhibits the interaction between ASC and the PYD domain of NLRP3, the inhibition of NF-κB p65 by POP2 is ASC-independent [123]. Additionally, COPs such as INCA, COP, and ICEBERG, as well as caspase-12, can inhibit the processing of pro-IL-1β and activation of caspase-1 by preventing the recruitment of ASC during inflammasome assembly [118–120]. Furthermore, NLRP12 can inhibit NF-κB and ERK signalling in dendritic cells and macrophages [124]. A previous report suggested that cellular K+ concentration plays an important role in regulating inflammasome activity since low amounts of K+ promote ASC assembly, resulting in inflammasome activation, whereas high concentrations of K+ block the secretion of cytokines [125]. This indicates that low intracellular K+ levels are required for the activation of inflammasomes [125]. However, some inflammasomes are more sensitive to high concentrations of K+ than others are and the mechanism behind this phenomenon is still elusive. Therefore, the mechanisms through which several inflammasomes triggers K+ efflux remain unclear [126].
Viral immune evasion mechanisms targeting NLRs and inflammasomes
To establish a productive infection, several viruses have developed evolutionary strategies enabling them to constantly adapt and evade the host immune response [127]. Several studies have identified certain specific mechanisms of viral evasion against the inflammasome, these may occur at different levels by (a) modifying the expression of viral molecules involved in detection by PRRs; (b) directly interacting with host proteins involved in the innate immune pathway; (c) inactivating effector molecules by binding to them; and (d) modulating the expression of inflammasome components [20]. Here, we discuss recent advances regarding the inflammasome evasion mechanisms employed by viruses (Fig. 4).
Viral strategies for preventing inflammasome activation. Inflammasome complex activation occurs through a two-step process, which is initiated by the stimulation of inflammasome components, leading to the production of pro-IL-1β and pro-IL-18 through NF-κB activation, followed by a final signal that triggers inflammasome complex activation resulting in caspase-1 catalysis. Viruses have developed several mechanisms to inhibit inflammasome assembly and activity. For example, preventing the translocation and transactivation of NF-κB would limit the synthesis of NLRP3 and the inflammasome substrates, IL-1β and IL-18. Among these viral mechanisms, the Orf63 protein of KSHV and the V protein of MV prevent inflammasome activation by inhibiting the oligomerisation of the NLRP1 and NLRP3 inflammasomes, respectively. Several Orthopoxvirus strains, including M013 myxoma virus and gp013L Shope fibroma virus, encode PYD-only proteins (POPs) that bind ASC to prevent caspase-1 recruitment. In addition, endogenous POPs such as POP1 and POP2 interact with the PYD domains of ASC and NLRs, respectively, and thus inhibit the activation of the inflammasome. Viral proteins can also inhibit inflammasome formation by hampering caspase-1 in different ways. Orthopoxvirus strains can also prevent the downstream effects of IL-1β/IL-18 via its binding proteins (BPs). In addition, the myxoma and vaccinia viruses encode serpins such as CrmA SP1/2 (SPI1/2) and Serp2 that can abolish the enzymatic activity of caspase-1. The NS1 protein of influenza virus prevents caspase-1 activation and the secretion of IL-1β and IL-18 via an unknown mechanism. The IL-1β and IL-18 pathways have been observed to be regulated by the vaccinia virus protein B15R and the molluscum contagiosum poxvirus proteins MC53L and MC54L, which can bind and inhibit IL-1β and IL-18, respectively
Regulation of the inflammasome
One mechanism that is used extensively by viruses to propagate their progeny is the regulation of host gene expression to control cellular factors involved in the infection cycle or inhibit defence mechanisms of the host [128, 129]. Several studies have suggested that viral factors participate in controlling the expression levels of inflammasome components. Data obtained from keratinocytes infected with papillomavirus demonstrate the strategies that have evolved to evade host immunity including transcriptional inhibition of the inflammasome component involving ASC, although the mechanisms remain to be elucidated [130]. Additionally, papilloma virus has been shown to possess a post-translational mechanism to regulate production of IL-1β; however, this process is regulated independently of NLRP3 [130]. Notably, this phenomenon is promoted by the E6 oncoprotein that activates the proteasome pathway and culminates in degradation of pro-IL-1β [131]. It is relevant to mention that many viral proteins can translocate into the nucleus of infected cells, probably to regulate the expression of genes involved in immunological processes [132, 133]. Thus, the viral alkaline exonuclease of Epstein Barr virus (BGLF5) down-regulates molecules of the immune system such as TLR-2 and CD1 by degrading the mRNAs that encode them [134]. The expression of inflammasome components is inhibited by a similar method during viral infections. Another viral strategy for immune evasion is the regulation of the expression of host genes by the production of post-transcriptional regulators such as microRNAs of both viral and cellular origin that can exert positive or negative regulation [135]. Viral miRNAs can directly alter components of the immune system. For example, γ-herpesviruses encode 30–40 miRNAs during infection and sequence analysis showed that two of them (miR-BART11-5P and miR-BART15) have a high homology with cellular miR-223, which can target NLRP3 and thereby restrict inflammasome activation in infected monocytes [136]. Further, in a study in which cells were cotransfected with a luciferase construct fused to the 3′ untranslated region of NLRP3 and one of several viral miRNAs, reduced luciferase activity was observed in the cells cotransfected with miR-BART15 [136].
Inhibition of inflammasome assembly
Viral genomes encode multifunctional proteins that possess diverse roles. Among these functions, the evasion of the immune response is an important strategy. Poxviruses and herpesviruses have large DNA genomes that encode various protein products, several of which can inhibit the assembly of the inflammasome to prevent its oligomerisation. Additionally, viral proteins can prevent the recruitment of ASC molecules, thus inhibiting the innate immune response and promoting a better environment for viral replication [137]. Numerous reports have shown that VACV encodes viral homologs of cellular proteins; for example, it encodes anti-apoptotic proteins with homology to the BCL-2 family that regulate the innate immune response by interacting with NLRP1 [138]. In detail, the F1L protein of VACV plays a key role in inhibiting the function of NLRP1, and this function depends on a specific region of F1L (residues 1–47). F1L has been suggested to interact with the binding region of ATP, which is required for NLRP1 oligomerisation. Due to this interaction, a reduction in the activity of caspase-1 and consequently, a decreased production of IL-1β has been observed, demonstrating that F1L acts as a viral NLRP1-suppressing factor [139]. Additionally, many viral proteins with amino acid sequences homologous to those of NLRP1 have been demonstrated, and of these viral proteins, many have homology with the LRR domain of NLRP1. Thus, the Orf63 protein of Kaposi’s sarcoma-associated herpesvirus (KSHV) is able to co-immunoprecipitate with NLRP1 protein. Furthermore, an analysis of the mutated domains in NLRP1 indicated that Orf63 protein interacts specifically with the LRR and NBD domains of NLRP1 and blocks the self-association of NLRP1 [140]. Measles virus V protein has been demonstrated to interact similarly with NLRP3 in infected cells. Further, it has been shown that cells infected with measles virus lacking the V gene show an increased production of IL-1β compared with those infected with the wild-type measles virus. These data support that the measles virus V protein inhibits inflammasome activation, although whether this occurs through NLRP3 oligomerisation remains unknown [48].
Besides, other viral proteins may inhibit inflammasome adaptor proteins to prevent the formation of a functional inflammasome complex, either by inhibiting their recruitment or preventing their oligomerisation [11]. In such cases, the virus must encode proteins with a PYD homology domain or homology to POPs. The significance of POPs in controlling host defence is underscored by the finding that some poxviruses encode viral POPs that block the activation of the inflammasome and NF-κB to escape the host immune response [141].
Another mechanism that viruses have evolved to evade the host immune response is to degrade the factors, receptors, and effector molecules involved in innate immune processes. This mechanism is underlined by a report that has shown that numerous viruses encode proteases that can act on immune molecules to inhibit inflammasome activation [142]. Recently, it has shown that the 2A and 3C proteases of enterovirus 71 can cleave NLRP3 at specific regions of its amino acid sequence (G493 or Q225-L494-G226); additionally, 3C protease interacts with NLRP3, culminating in the inhibition of inflammasome activation [143].
Inhibition of caspase-1
After inflammasome activation, major remaining events include the secretion of active IL-1β and activation of caspase-1. Some viruses have developed strategies to escape from the host immune response, including preventing the secretion of IL-1β. Thus, some viruses encode proteins that target caspase-1. Data regarding such viral mechanisms have been accumulating, with reports that viruses such as VACV, myxoma virus, and cowpox viruses encode viral proteins with serpin activity that acts on IL-1β-converting enzyme [144–146]. Viral proteins, CrmA, B13R, and Serp2, have been shown to have inhibitory effects on the activity of caspase-1, leading to the inhibition of the processing of IL-1β [144–146]. Another viral protein involved in this mechanism of immune evasion is the NS1 protein of influenza virus. An early report demonstrated that cells infected with wild-type influenza virus showed reduced IL-1β secretion compared with those infected with influenza virus mutated in the NS1 gene region. Further treatment with inhibitors of caspase-1 was found to significantly downregulate the production of this cytokine; however, the direct interaction between NS1 and caspase-1 is still elusive [147]. Recently, it was determined that NS1 physically interacts with endogenous NLRP3 and consequently abrogates the activation of NLRP3 in NS1-expressing THP-1 cells.
Furthermore, NS1 was found to inhibit the transactivation of the nuclear NF-κB, a major transcriptional activator, leading to impairment of the transcription of proinflammatory cytokines. These findings suggest that influenza virus NS1 protein targets both the NLRP3 and NF-κB, thus abrogating inflammasome activation, as a viral immune evasion strategy [148]. In conclusion, current evidence of new viral proteins that regulate the innate immune response specifically with regard to inflammasome activation is a critical issue that is gaining momentum. Understanding each of these events will be of great importance for the future development of new antiviral strategies against several viral infections.
Conclusions
During the past several years, inflammasome research has demonstrated the importance of NLR and non-NLR inflammasomes as pathogen sensors in the context of cellular recognition of viruses. The above information demonstrates the significance of NLR proteins and the inflammasome in innate immune signalling and host defence. Inflammasome activation plays an essential role in the immune response to viral infection and represents a potential target for drug- or vaccine-based interventions for the control of excessive inflammasome activation and immunopathology or for the control of viral replication. However, our knowledge of inflammasome biology with respect to viral infection is still at an early stage, and the complex mechanisms of inflammasome activation and its subsequent involvement in antiviral immunity are incompletely understood. Investigation over the past decades has elucidated several viral strategies for evading host immune responses, and the study of these processes has enhanced our mechanistic understanding of viral infection and its pathogenesis as well as revealed several targets of viral proteins in host cellular pathways. Thus, a more detailed analysis of viral immune evasion molecules that affect inflammasome function can be expected to unravel new biological processes and may ultimately lead to the identification of targets for the management and prevention of infectious diseases.
References
Medzhitov R, Janeway CA Jr. Innate immunity: impact on the adaptive immune response. Curr Opin Immunol. 1997;9(1):4–9.
Medzhitov R, Janeway CA Jr. Decoding the patterns of self and nonself by the innate immune system. Science (80-). 2002;296(5566):298–300. doi:10.1126/science.1068883.
Wilkins C, Gale M. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22(1):41–7. doi:10.1016/j.coi.2009.12.003.
Pothlichet J, Meunier I, Davis BK, et al. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog. 2013;9(4):e1003256. doi:10.1371/journal.ppat.1003256.
Lawrence TM, Hudacek AW, de Zoete MR, Flavell RA, Schnell MJ. Rabies virus is recognized by the NLRP3 inflammasome and activates interleukin-1β release in murine dendritic cells. J Virol. 2013;87(10):5848–57. doi:10.1128/JVI.00203-13.
Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153(2):348–61. doi:10.1016/j.cell.2013.02.054.
Park S, Juliana C, Hong S, et al. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J Immunol. 2013;191(8):4358–66. doi:10.4049/jimmunol.1301170.
Cai X, Chen J, Xu H, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156(6):1207–22. doi:10.1016/j.cell.2014.01.063.
Franchi L, Eigenbrod T, Muñoz-Planillo R, et al. Cytosolic double-stranded RNA activates the NLRP3 inflammasome via MAVS-induced membrane permeabilization and K+ efflux. J Immunol. 2014;193(8):4214–22. doi:10.4049/jimmunol.1400582.
Huang H, Saravia J, You D, Shaw AJ, Cormier SA. Impaired gamma delta T cell-derived IL-17A and inflammasome activation during early respiratory syncytial virus infection in infants. Immunol Cell Biol. 2014;. doi:10.1038/icb.2014.79.
Lupfer C, Malik A, Kanneganti T-D. Inflammasome control of viral infection. Curr Opin Virol. 2015;12:38–46. doi:10.1016/j.coviro.2015.02.007.
Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39(6):1003–18. doi:10.1016/j.immuni.2013.11.010.
Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1beta secretion. Cytokine Growth Factor Rev. 2011;22(4):189–95. doi:10.1016/j.cytogfr.2011.10.001.
Rietdijk ST, Burwell T, Bertin J, Coyle AJ. Sensing intracellular pathogens-NOD-like receptors. Curr Opin Pharmacol. 2008;8(3):261–6. doi:10.1016/j.coph.2008.04.003.
Lamkanfi M, Kanneganti TD. Nlrp3: an immune sensor of cellular stress and infection. Int J Biochem Cell Biol. 2010;42(6):792–5. doi:10.1016/j.biocel.2010.01.008.
Hornung V, Ablasser A, Charrel-Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–8. doi:10.1038/nature07725.
Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10(2):417–26.
Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–22. doi:10.1016/j.cell.2014.04.007.
Cridland JA, Curley EZ, Wykes MN, et al. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12(1):140. doi:10.1186/1471-2148-12-140.
Gram AM, Frenkel J, Ressing ME. Inflammasomes and viruses: cellular defence versus viral offence. J Gen Virol. 2012;93(Pt 10):2063–75. doi:10.1099/vir.0.042978-0.
Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442(7098):39–44. doi:10.1038/nature04946.
Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7(12):1250–7. doi:10.1038/ni1412.
Werts C, Girardin SE, Philpott DJ. TIR, CARD and PYRIN: three domains for an antimicrobial triad. Cell Death Differ. 2006;13(5):798–815. doi:10.1038/sj.cdd.4401890.
Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3(5):371–82. doi:10.1038/nri1086.
Mariathasan S, Newton K, Monack DM, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–8. doi:10.1038/nature02664.
Faustin B, Reed JC. Sunburned skin activates inflammasomes. Trends Cell Biol. 2008;18(1):4–8. doi:10.1016/j.tcb.2007.10.004.
Tattoli I, Travassos LH, Carneiro LA, Magalhaes JG, Girardin SE. The Nodosome: Nod1 and Nod2 control bacterial infections and inflammation. Semin Immunopathol. 2007;29(3):289–301. doi:10.1007/s00281-007-0083-2.
Lu C, Wang A, Wang L, Dorsch M, Ocain TD, Xu Y. Nucleotide binding to CARD12 and its role in CARD12-mediated caspase-1 activation. Biochem Biophys Res Commun. 2005;331(4):1114–9. doi:10.1016/j.bbrc.2005.04.027.
Lupfer C, Kanneganti T-D. The expanding role of NLRs in antiviral immunity. Immunol Rev. 2013;255(1):13–24. doi:10.1111/imr.12089.
Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol Rev. 2011;243(1):136–51.
Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14(1):10–22. doi:10.1038/sj.cdd.4402038.
Bauernfeind FG, Horvath G, Stutz A, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183(2):787–91. doi:10.4049/jimmunol.0901363.
Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27(1):229–65. doi:10.1146/annurev.immunol.021908.132715.
Hornung V, Bauernfeind F, Halle A, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9(8):847–56. doi:10.1038/ni.1631.
Cruz CM, Rinna A, Forman HJ, Ventura AL, Persechini PM, Ojcius DM. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282(5):2871–9. doi:10.1074/jbc.M608083200.
Kanneganti TD, Lamkanfi M, Kim YG, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26(4):433–43. doi:10.1016/j.immuni.2007.03.008.
Murakami T, Ockinger J, Yu J, et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci USA. 2012;109(28):11282–7. doi:10.1073/pnas.1117765109.
Lee G-S, Subramanian N, Kim AI, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492(7427):123–7. doi:10.1038/nature11588.
Feldmeyer L, Keller M, Niklaus G, Hohl D, Werner S, Beer HD. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr Biol. 2007;17(13):1140–5. doi:10.1016/j.cub.2007.05.074.
Triantafilou K, Hughes TR, Triantafilou M, Morgan BP. The complement membrane attack complex triggers intracellular Ca2+ fluxes leading to NLRP3 inflammasome activation. J Cell Sci. 2013;126(Pt 13):2903–13. doi:10.1242/jcs.124388.
Rossol M, Pierer M, Raulien N, et al. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nat Commun. 2012;3:1329.
Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38(6):1142–53. doi:10.1016/j.immuni.2013.05.016.
Guo HC, Jin Y, Zhi XY, Yan D, Sun SQ. NLRP3 inflammasome activation by viroporins of animal viruses. Viruses. 2015;7(7):3380–91. doi:10.3390/v7072777.
Chen IY, Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23(1):55–63. doi:10.1016/j.tim.2014.09.007.
Kanneganti TD, Body-Malapel M, Amer A, et al. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281(48):36560–8. doi:10.1074/jbc.M607594200.
Rajan JV, Rodriguez D, Miao EA, Aderem A. The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. J Virol. 2011;85(9):4167–72. doi:10.1128/JVI.01687-10.
Ito M, Yanagi Y, Ichinohe T. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog. 2012;8(8):e1002857. doi:10.1371/journal.ppat.1002857.
Komune N, Ichinohe T, Ito M, Yanagi Y. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1 secretion. J Virol. 2011;85(24):13019–26. doi:10.1128/JVI.05942-11.
Triantafilou K, Kar S, Van Kuppeveld FJM, Triantafilou M. Rhinovirus-induced calcium flux triggers NLRP3 and NLRC5 activation in bronchial cells. Am J Respir Cell Mol Biol. 2013;49:923–34. doi:10.1165/rcmb.2013-0032OC.
Shrivastava S, Mukherjee A, Ray R, Ray RB. Hepatitis C virus induces interleukin-1 (IL-1)/IL-18 in circulatory and resident liver macrophages. J Virol. 2013;87(22):12284–90. doi:10.1128/JVI.01962-13.
Negash AA, Ramos HJ, Crochet N, et al. IL-1β production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog. 2013;9(4):e1003330. doi:10.1371/journal.ppat.1003330.
Chen W, Xu Y, Li H, et al. HCV genomic RNA activates the NLRP3 inflammasome in human myeloid cells. PLoS ONE. 2014;9(1):e84953. doi:10.1371/journal.pone.0084953.
Owen DM, Gale M. Fighting the flu with inflammasome signaling. Immunity. 2009;30(4):476–8. doi:10.1016/j.immuni.2009.03.011.
Pang IK, Iwasaki A. Inflammasomes as mediators of immunity against influenza virus. Trends Immunol. 2011;32(1):34–41. doi:10.1016/j.it.2010.11.004.
Allen IC, Scull MA, Moore CB, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–65. doi:10.1016/j.immuni.2009.02.005.
Thomas PG, Dash P, Aldridge JR Jr, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30(4):566–75. doi:10.1016/j.immuni.2009.02.006.
Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11(5):404–10. doi:10.1038/ni.1861.
Segovia J, Sabbah A, Mgbemena V, et al. TLR2/MyD88/NF-kappaB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. PLoS ONE. 2012;7(1):e29695. doi:10.1371/journal.pone.0029695.
McAuley JL, Tate MD, MacKenzie-Kludas CJ, et al. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 2013;. doi:10.1371/journal.ppat.1003392.
Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax. 2013;68(1):66–75. doi:10.1136/thoraxjnl-2012-202182.
Pontillo A, Silva LT, Oshiro TM, Finazzo C, Crovella S, Duarte AJS. HIV-1 induces NALP3-inflammasome expression and interleukin-1β secretion in dendritic cells from healthy individuals but not from HIV-positive patients. AIDS. 2012;26(1):11–8. doi:10.1097/QAD.0b013e32834d697f.
Ramos HJ, Lanteri MC, Blahnik G, et al. IL-1β signaling promotes CNS-intrinsic immune control of West Nile virus infection. PLoS Pathog. 2012;8(11):e1003039. doi:10.1371/journal.ppat.1003039.
Kumar M, Roe K, Orillo B, et al. Inflammasome adaptor protein Apoptosis-associated speck-like protein containing CARD (ASC) is critical for the immune response and survival in west Nile virus encephalitis. J Virol. 2013;87(7):3655–67. doi:10.1128/JVI.02667-12.
Kaushik DK, Gupta M, Kumawat KL, Basu A. NLRP3 inflammasome: key mediator of neuroinflammation in murine Japanese encephalitis. PLoS ONE. 2012;7(2):e32270. doi:10.1371/journal.pone.0032270.
Hottz ED, Lopes JF, Freitas C, et al. Platelets mediate increased endothelium permeability in dengue through NLRP3-inflammasome activation. Blood. 2013;122(20):3405–14. doi:10.1182/blood-2013-05-504449.
Wu M-F, Chen S-T, Yang A-H, et al. CLEC5A is critical for dengue virus-induced inflammasome activation in human macrophages. Blood. 2013;121(1):95–106. doi:10.1182/blood-2012-05-430090.
Ermler ME, Traylor Z, Patel K, et al. Rift valley fever virus infection induces activation of the NLRP3 inflammasome. Virology. 2014;449:174–80. doi:10.1016/j.virol.2013.11.015.
Wikan N, Khongwichit S, Phuklia W, et al. Comprehensive proteomic analysis of white blood cells from chikungunya fever patients of different severities. J Transl Med. 2014;12(1):96. doi:10.1186/1479-5876-12-96.
Muruve DA, Pétrilli V, Zaiss AK, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452(7183):103–7. doi:10.1038/nature06664.
Delaloye J, Roger T, Steiner-Tardivel Q-G, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009;5(6):e1000480. doi:10.1371/journal.ppat.1000480.
Di Paolo NC, Miao EA, Iwakura Y, et al. Virus binding to a plasma membrane receptor triggers interleukin-1α-mediated proinflammatory macrophage response in vivo. Immunity. 2009;31(1):110–21. doi:10.1016/j.immuni.2009.04.015.
Barlan AU, Danthi P, Wiethoff CM. Lysosomal localization and mechanism of membrane penetration influence nonenveloped virus activation of the NLRP3 inflammasome. Virology. 2011;412(2):306–14. doi:10.1016/j.virol.2011.01.019.
Barlan AU, Griffin TM, McGuire KA, Wiethoff CM. Adenovirus membrane penetration activates the NLRP3 inflammasome. J Virol. 2011;85(1):146–55. doi:10.1128/JVI.01265-10.
Nour AM, Reichelt M, Ku C-C, Ho M-Y, Heineman TC, Arvin AM. Varicella-zoster virus infection triggers formation of an interleukin-1β (IL-1β)-processing inflammasome complex. J Biol Chem. 2011;286(20):17921–33. doi:10.1074/jbc.M110.210575.
Roberts TL, Idris A, Dunn JA, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–60. doi:10.1126/science.1169841.
Burckstummer T, Baumann C, Bluml S, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10(3):266–72. doi:10.1038/ni.1702.
Rathinam VA, Jiang Z, Waggoner SN, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11(5):395–402. doi:10.1038/ni.1864.
Ekchariyawat P, Hamel R, Bernard E, et al. Inflammasome signaling pathways exert antiviral effect against Chikungunya virus in human dermal fibroblasts. Infect Genet Evol. 2015;32:401–8. doi:10.1016/j.meegid.2015.03.025.
Hamel R, Dejarnac O, Wichit S, et al. Biology of Zika virus infection in human skin cells. J Virol. 2015;. doi:10.1128/JVI.00354-15.
Kawai T, Takahashi K, Sato S, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–8. doi:10.1038/ni1243.
Rehwinkel J, Tan CP, Goubau D, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140(3):397–408. doi:10.1016/j.cell.2010.01.020.
Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–7. doi:10.1038/ni1087.
Kolakofsky D, Kowalinski E, Cusack S. A structure-based model of RIG-I activation. RNA. 2012;18(12):2118–27. doi:10.1261/rna.035949.112.
Hornung V, Ellegast J, Kim S, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science (80-). 2006;314(5801):994–7. doi:10.1126/science.1132505.
Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–5. doi:10.1038/nature04734.
Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol. 2007;81(3):1401–11. doi:10.1128/JVI.01740-06.
Fredericksen BL, Gale M. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80(6):2913–23. doi:10.1128/JVI.80.6.2913-2923.2006.
Melchjorsen J, Jensen SB, Malmgaard L, et al. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol. 2005;79(20):12944–51. doi:10.1128/JVI.79.20.12944-12951.2005.
Mikkelsen SS, Jensen SB, Chiliveru S, et al. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells. Dependence on TRAF2 and TAK1. J Biol Chem. 2009;284(16):10774–82. doi:10.1074/jbc.M807272200.
Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science (80-). 2006;314(5801):997–1001. doi:10.1126/science.1132998.
Rothenfusser S, Goutagny N, DiPerna G, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175(8):5260–8. doi:10.4049/jimmunol.175.8.5260.
Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82(2):609–16. doi:10.1128/JVI.01305-07.
Chang TH, Liao CL, Lin YL. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3 K-dependent NF-κB activation. Microbes Infect. 2006;8:157–71. doi:10.1016/j.micinf.2005.06.014.
Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(July):523–7. doi:10.1038/nature07106.
Poeck H, Bscheider M, Gross O, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11(1):63–9. doi:10.1038/ni.1824.
Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185(3):1681–91. doi:10.4049/jimmunol.0903900.
Cui J, Zhu L, Xia X, et al. NLRC5 Negatively regulates the NF-κB and type I interferon signaling pathways. Cell. 2010;141(3):483–96. doi:10.1016/j.cell.2010.03.040.
Kuenzel S, Till A, Winkler M, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184(4):1990–2000. doi:10.4049/jimmunol.0900557.
Meissner TB, Li A, Biswas A, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA. 2010;107(31):13794–9. doi:10.1073/pnas.1008684107.
Neerincx A, Lautz K, Menning M, et al. A role for the human NLR family member NLRC5 in antiviral responses. J Biol Chem. 2010;285(34):26223–32. doi:10.1074/jbc.M110.109736.
Davis BK, Roberts RA, Huang MT, et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2011;186(3):1333–7. doi:10.4049/jimmunol.1003111.
Moore CB, Bergstralh DT, Duncan JA, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451(7178):573–7.
Tattoli I, Carneiro LA, Jéhanno M, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi:10.1038/sj.embor.7401161.
Arnoult D, Soares F, Tattoli I, Castanier C, Philpott DJ, Girardin SE. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122(Pt 17):3161–8.
Lei Y, Wen H, Yu Y, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36(6):933–46.
Lei Y, Wen H, Ting JPY. The NLR protein, NLRX1, and its partner, TUFM, reduce type I interferon, and enhance autophagy. Autophagy. 2013;9(3):432–3.
Jaworska J, Coulombe F, Downey J, et al. NLRX1 prevents mitochondrial induced apoptosis and enhances macrophage antiviral immunity by interacting with influenza virus PB1-F2 protein. Proc Natl Acad Sci USA. 2014;. doi:10.1073/pnas.0507681102.
Unger BL, Ganesan S, Comstock AT, Faris AN, Hershenson MB, Sajjan US. Nod-like receptor X-1 is required for rhinovirus-induced barrier dysfunction in airway epithelial cells. J Virol. 2014;88(7):3705–3718. http://www.ncbi.nlm.nih.gov/pubmed/24429360.
Guo H, König R, Deng M, et al. NLRX1 sequesters STING to negatively regulate the interferon response, thereby facilitating the replication of HIV-1 and DNA viruses. Cell Host Microbe. 2016;19(4):515–28.
Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev. 2015;265(1):130–42. doi:10.1111/imr.12287.
Fink SL, Cookson BT. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006;8(11):1812–25.
Denes A, Lopez-Castejon G, Brough D. Caspase-1: is IL-1 just the tip of the ICEberg? Cell Death Dis. 2012;3(7):e338. doi:10.1038/cddis.2012.86.
Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243(1):206–14. doi:10.1111/j.1600-065X.2011.01044.x.
Asgari E, Le Friec G, Yamamoto H, et al. C3a modulates IL-1β secretion in human monocytes by regulating ATP efflux and subsequent NLRP3 inflammasome activation. Blood. 2013;122(20):3473–81. doi:10.1182/blood-2013-05-502229.
de Gassart A, Martinon F. Pyroptosis: caspase-11 unlocks the gates of death. Immunity. 2015;43(5):835–7.
Yang D, He Y, Muñoz-Planillo R, Liu Q, Núñez G. Caspase-11 requires the pannexin-1 channel and the purinergic P2X7 pore to mediate pyroptosis and endotoxic shock. Immunity. 2015;. doi:10.1016/j.immuni.2015.10.009.
Chekeni FB, Elliott MR, Sandilos JK, et al. Pannexin 1 channels mediate “find-me” signal release and membrane permeability during apoptosis. Nature. 2010;467(7317):863–867. http://www.nature.com.iclibezp1.cc.ic.ac.uk/nature/journal/v467/n7317/full/nature09413.html.
Lu A, Magupalli VG, Ruan J, et al. Unified polymerization mechanism for the assembly of asc-dependent inflammasomes. Cell. 2014;156(6):1193–206. doi:10.1016/j.cell.2014.02.008.
Lu A, Wu H. Structural mechanisms of inflammasome assembly. FEBS J. 2015;282(3):435–44. doi:10.1111/febs.13133.
Le HT, Harton JA. Pyrin- and CARD-only proteins as regulators of NLR functions. Front Immunol. 2013;4:275. doi:10.3389/fimmu.2013.00275.
Stehlik C, Fiorentino L, Dorfleutner A, et al. The PAAD/PYRIN-family protein ASC is a dual regulator of a conserved step in nuclear factor B activation pathways. J Exp Med. 2002;196(12):1605–15. doi:10.1084/jem.20021552.
Bedoya F, Sandler LL, Harton JA. Pyrin-only protein 2 modulates NF-κB and disrupts ASC: CLR interactions. J Immunol. 2007;178(6):3837–45. doi:10.4049/jimmunol.178.6.3837.
Atianand MK, Harton JA. Uncoupling of pyrin-only protein 2 (POP2)-mediated dual regulation of NF-κB and the inflammasome. J Biol Chem. 2011;286(47):40536–47. doi:10.1074/jbc.M111.274290.
Zamoshnikova A, Groß CJ, Schuster S, et al. NLRP12 is a neutrophil-specific, negative regulator of in vitro cell migration but does not modulate LPS- or infection-induced NF-κB or ERK signalling. Immunobiology. 2016;221(2):341–6. doi:10.1016/j.imbio.2015.10.001.
Khare S, Ratsimandresy RA, de Almeida L, et al. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol. 2014;15(4):343–353. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4123781&tool=pmcentrez&rendertype=abstract.
Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. doi:10.1038/nri3452.
Devasthanam AS. Mechanisms underlying the inhibition of interferon signaling by viruses. Virulence. 2014;5(2):270–7. doi:10.4161/viru.27902.
Gan CS, Yusof R, Othman S. Different serotypes of dengue viruses differently regulate the expression of the host cell antigen processing machinery. Acta Trop. 2015;149:8–14. doi:10.1016/j.actatropica.2015.05.005.
Yang Q, Fu S, Wang J. Hepatitis C virus infection decreases the expression of Toll-like receptors 3 and 7 via upregulation of miR-758. Arch Virol. 2014. doi:10.1007/s00705-014-2167-3.
Karim R, Meyers C, Backendorf C, et al. Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PLoS ONE. 2011;6(3):e17848. doi:10.1371/journal.pone.0017848.
Niebler M, Qian X, Höfler D, et al. Post-translational control of IL-1b via the human papillomavirus type 16 E6 oncoprotein: a novel mechanism of innate immune escape mediated by the E3-ubiquitin ligase E6-AP and p53. PLoS Pathog. 2013;9(8):e1003536. doi:10.1371/journal.ppat.1003536.
Radko S, Koleva M, James KMD, Jung R, Mymryk JS, Pelka P. Adenovirus E1A targets the DREF nuclear factor to regulate virus gene expression, DNA replication, and growth. J Virol. 2014;88(22):13469–81. doi:10.1128/JVI.02538-14.
Wulan WN, Heydet D, Walker EJ, Gahan ME, Ghildyal R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front Microbiol. 2015;6:1–10. doi:10.3389/fmicb.2015.00553.
Rowe M, Glaunsinger B, van Leeuwen D, et al. Host shutoff during productive Epstein–Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci USA. 2007;104(9):3366–71. doi:10.1073/pnas.0611128104.
Tycowski KT, Guo YE, Lee N, et al. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29(6):567–84. doi:10.1101/gad.259077.115.
Haneklaus M, Gerlic M, Kurowska-Stolarska M, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1 production. J Immunol. 2012;189(8):3795–9. doi:10.4049/jimmunol.1200312.
Lupfer CR, Kanneganti T-D. The role of inflammasome modulation in virulence. Virulence. 2012;3(3):262–70. doi:10.4161/viru.20266.
Marshall B, Puthalakath H, Caria S, et al. Variola virus F1L is a Bcl-2-like protein that unlike its vaccinia virus counterpart inhibits apoptosis independent of Bim. Cell Death Dis. 2015;6(3):e1680. doi:10.1038/cddis.2015.52.
Gerlic M, Faustin B, Postigo A, et al. Vaccinia virus F1L protein promotes virulence by inhibiting inflammasome activation. Proc Natl Acad Sci USA. 2013;110(19):7808–13. doi:10.1073/pnas.1215995110.
Gregory SM, Davis BK, West JA, et al. Discovery of a viral NLR homolog that inhibits the inflammasome. Science (80-). 2011;331(6015):330–4. doi:10.1126/science.1199478.
Johnston JB, Barrett JW, Nazarian SH, et al. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23(6):587–98. doi:10.1016/j.immuni.2005.10.003.
Wang D, Fang L, Wei D, et al. Hepatitis A virus 3C protease cleaves NEMO to impair induction of beta interferon. J Virol. 2014;88(17):10252–8. doi:10.1128/JVI.00869-14.
Wang H, Lei X, Xiao X, et al. Reciprocal regulation between enterovirus 71 and the NLRP3 inflammasome. Cell Rep. 2015;12(1):1–7. doi:10.1016/j.celrep.2015.05.047.
Ray CA, Black RA, Kronheim SR, et al. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69(4):597–604. doi:10.1016/0092-8674(92)90223-Y.
Petit F, Bertagnoli S, Gelfi J, Fassy F, Boucraut-Baralon C, Milon A. Characterization of a myxoma virus-encoded serpin-like protein with activity against interleukin-1 beta-converting enzyme. J Virol. 1996;70(9):5860–5866. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8709205\nhttp://jvi.asm.org/content/70/9/5860.full.pdf.
Kettle S, Alcamí A, Khanna A, Ehret R, Jassoy C, Smith GL. Vaccinia virus serpin B13R (SPI-2) inhibits interleukin-1beta-converting enzyme and protects virus-infected cells from TNF- and Fas-mediated apoptosis, but does not prevent IL-1beta-induced fever. J Gen Virol. 1997;78(Pt 3):677–85.
Stasakova J. Influenza A mutant viruses with altered NS1 protein function provoke caspase-1 activation in primary human macrophages, resulting in fast apoptosis and release of high levels of interleukins 1 and 18. J Gen Virol. 2005;86(1):185–95. doi:10.1099/vir.0.80422-0.
Cheong W-C, Kang H-R, Yoon H, Kang S-J, Ting JP-Y, Song MJ. Influenza A virus NS1 protein inhibits the NLRP3 inflammasome. PLoS ONE. 2015;10(5):e0126456. doi:10.1371/journal.pone.0126456.
Acknowledgments
We would like to thank all laboratory members (Sofía Carrillo Halfon, Paola Carolina Valenzuela León, Josselin Carolina Corzo Gómez, and Alfredo Montes Gómez) for their constant help and support.
Funding
This study was supported by the National Council for Science and Technology (CONACyT, Grant Salud-2014-1-233347). GS received fellowships from CONACyT; LC-B, ML-J and JG-C are members of the National System of Researchers. The funding agencies were not involved in the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Rights and permissions
About this article
Cite this article
Shrivastava, G., León-Juárez, M., García-Cordero, J. et al. Inflammasomes and its importance in viral infections. Immunol Res 64, 1101–1117 (2016). https://doi.org/10.1007/s12026-016-8873-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-016-8873-z