Abstract
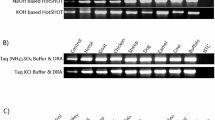
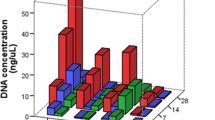
The breakdown of DNA and RNA in decomposing human tissue represents a major obstacle for postmortem forensic molecular analysis. This study investigated the feasibility of performing PCR-based molecular analysis of blood and muscle tissue from 45 autopsy cases with defined postmortem intervals ranging from one to more than 14 days. It was not possible to collect blood from 38 % of the autopsy cases due to severe coagulation and hemolysis, whereas muscle tissue was available for all cases. PCR-amplifiable DNA could be extracted from 96 % of the frozen muscle specimens and from 93 % of the formalin fixed and paraffin embedded (FFPE) muscle specimens. A quality assessment of muscle-derived DNA showed increased fragmentation with advancing body decomposition and generally more fragmentation in DNA from FFPE tissue than in DNA from frozen tissue. It was possible to amplify 1,000 basepair (bp) DNA fragments from all samples with postmortem intervals below 3 days whereas 400–600 bp long fragments typically could be amplified from the most decomposed muscle specimens. RNA was less stable than DNA in postmortem muscle tissue, yet selected mRNA molecules could be detected by reverse-transcriptase PCR in all samples up to 3 days after death. We conclude that analysis of DNA from bodies with a wide postmortem interval range is usually possible whereas the consistency of RNA analyses decreases considerably 3 days postmortem. We showed that muscle tissue is a highly usable source of DNA and RNA for postmortem forensic molecular analysis as well as for retrospective research projects based on archived FFPE specimens.

Similar content being viewed by others
References
Madea B, Saukko P, Oliva A, Musshoff F. Molecular pathology in forensic medicine-Introduction. Forensic Sci Int. 2010;203:3–14.
Tester DJ, Ackerman MJ. The role of molecular autopsy in unexplained sudden cardiac death. Curr Opin Cardiol. 2006;21:166–72.
Winkel BG, Larsen MK, Berge KE, Leren TP, Nissen PH, Olesen MS, et al. The prevalence of mutations in KCNQ1, KCNH2, and SCN5A in an unselected national cohort of young sudden unexplained death cases. J Cardiovasc Electrophysiol. 2012;23:1092–8.
Basso C, Carturan E, Pilichou K, Rizzo S, Corrado D, Thiene G. Sudden cardiac death with normal heart: molecular autopsy. Cardiovasc Pathol. 2010;19:321–5.
Di PM, Luchini D, Bloise R, Priori SG. Postmortem molecular analysis in victims of sudden unexplained death. Am J Forensic Med Pathol. 2004;25:182–4.
Gaaloul I, Riabi S, Harrath R, Evans M, Salem NH, Mlayeh S, Huber S, Aouni M. Sudden unexpected death related to enterovirus myocarditis: histopathology, immunohistochemistry and molecular pathology diagnosis at post-mortem. BMC Infect Dis. 2013;12:212.
Feldman MY. Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog Nucleic Acid Res Mol Biol. 1973;13:1–49.
Bonin S, Petrera F, Niccolini B, Stanta G. PCR analysis in archival postmortem tissues. Mol Pathol. 2003;56:184–6.
Bonin S, Hlubek F, Benhattar J, Denkert C, Dietel M, Fernandez PL, et al. Multicentre validation study of nucleic acids extraction from FFPE tissues. Virchows Arch. 2010;457:309–17.
Gillio-Tos A, De ML, Fiano V, Garcia-Bragado F, Dikshit R, Boffetta P, Merletti F. Efficient DNA extraction from 25-year-old paraffin-embedded tissues: study of 365 samples. Pathology. 2007;39:345–8.
van Beers EH, Joosse SA, Ligtenberg MJ, Fles R, Hogervorst FB, Verhoef S, Nederlof PM. A multiplex PCR predictor for aCGH success of FFPE samples. Br J Cancer. 2006;94:333–7.
von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS One. 2007;2:e1261.
Huijsmans CJ, Damen J, van der Linden JC, Savelkoul PH, Hermans MH. Comparative analysis of four methods to extract DNA from paraffin-embedded tissues: effect on downstream molecular applications. BMC Res Notes. 2010;3:239.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86.
Sikora MJ, Thibert JN, Salter J, Dowsett M, Johnson MD, Rae JM. High-efficiency genotype analysis from formalin-fixed, paraffin-embedded tumor tissues. Pharmacogenomic J. 2011;11(5):348–58.
Hansen J, Corydon TJ, Palmfeldt J, Durr A, Fontaine B, Nielsen MN, Christensen JH, Gregersen N, Bross P. Decreased expression of the mitochondrial matrix proteases Lon and ClpP in cells from a patient with hereditary spastic paraplegia (SPG13). Neuroscience. 2008;153:474–82.
Bar W, Kratzer A, Machler M, Schmid W. Postmortem stability of DNA. Forensic Sci Int. 2008;39:59–70.
Ludes B, Pfitzinger H, Mangin P. DNA fingerprinting from tissues after variable postmortem periods. J Forensic Sci. 1993;38:686–90.
Sato Y, Motani H, Inoue H, Hayakawa M, Yajima D, Nagasawa S, et al. Multiplex STR typing of aortic tissues from unidentified cadavers. Leg Med (Tokyo). 2009;11(Suppl 1):S455–7.
Schwark T, Heinrich A, von Wurmb-Schwark N. Genetic identification of highly putrefied bodies using DNA from soft tissues. Int J Legal Med. 2011;125(6):891–4.
Kosel S, Grasbon-Frodl EM, Arima K, Chimelli L, Hahn M, Hashizume Y, et al. Inter-laboratory comparison of DNA preservation in archival paraffin-embedded human brain tissue from participating centres on four continents. Neurogenetics. 2001;3:163–70.
Farrugia A, Keyser C, Ludes B. Efficiency evaluation of a DNA extraction and purification protocol on archival formalin-fixed and paraffin-embedded tissue. Forensic Sci Int. 2010;194:e25–8.
Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van ME, Worobey M. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2(6):e537.
Vennemann M, Koppelkamm A. mRNA profiling in forensic genetics I: possibilities and limitations. Forensic Sci Int. 2010;203:71–5.
Johnson SA, Morgan DG, Finch CE. Extensive postmortem stability of RNA from rat and human brain. J Neurosci Res. 1986;16:267–80.
Schramm M, Falkai P, Tepest R, Schneider-Axmann T, Przkora R, Waha A, et al. Stability of RNA transcripts in post-mortem psychiatric brains. J Neural Transm. 1999;106:329–35.
Bauer M, Gramlich I, Polzin S, Patzelt D. Quantification of mRNA degradation as possible indicator of postmortem interval-a pilot study. Leg Med (Tokyo). 2003;5:220–7.
Preece P, Cairns NJ. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Res Mol Brain Res. 2003;118:60–71.
Partemi S, Berne PM, Batlle M, Berruezo A, Mont L, Riuro H, et al. Analysis of mRNA from human heart tissue and putative applications in forensic molecular pathology. Forensic Sci Int. 2010;203:99–105.
Heinrich M, Matt K, Lutz-Bonengel S, Schmidt U. Successful RNA extraction from various human postmortem tissues. Int J Legal Med. 2007;121:136–42.
Inoue H, Kimura A, Tuji T. Degradation profile of mRNA in a dead rat body: basic semi-quantification study. Forensic Sci Int. 2002;130:127–32.
Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neurosci Lett. 1995;200:151–4.
Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–43.
Graham EA, Turk EE, Rutty GN. Room temperature DNA preservation of soft tissue for rapid DNA extraction: an addition to the disaster victim identification investigators toolkit? Forensic Sci Int Genet. 2008;2:29–34.
Caputo M, Bosio LA, Corach D. Long-term room temperature preservation of corpse soft tissue: an approach for tissue sample storage. Investig Genet. 2011;2:7.
Butler JM, Shen Y, McCord BR. The development of reduced size STR amplicons as tools for analysis of degraded DNA. J Forensic Sci. 2003;48:1054–64.
Senge T, Madea B, Junge A, Rothschild MA, Schneider PM. STRs, mini STRs and SNPs—a comparative study for typing degraded DNA. Leg Med (Tokyo). 2011;13:68–74.
von Wurmb-Schwark N, Preusse-Prange A, Heinrich A, Simeoni E, Bosch T, Schwark T. A new multiplex-PCR comprising autosomal and y-specific STRs and mitochondrial DNA to analyze highly degraded material. Forensic Sci Int Genet. 2009;3:96–103.
Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH. Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5′ nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn. 2000;2:84–91.
Abrahamsen HN, Steiniche T, Nexo E, Hamilton-Dutoit SJ, Sorensen BS. Towards quantitative mRNA analysis in paraffin-embedded tissues using real-time reverse transcriptase-polymerase chain reaction: a methodological study on lymph nodes from melanoma patients. J Mol Diagn. 2003;5:34–41.
Oberli A, Popovici V, Delorenzi M, Baltzer A, Antonov J, Matthey S, et al. Expression profiling with RNA from formalin-fixed, paraffin-embedded material. BMC Med Genomics. 2008;1:9.
Kondo T, Ishida Y. Molecular pathology of wound healing. Forensic Sci Int. 2010;203:93–8.
Bauer M. RNA in forensic science. Forensic Sci Int Genet. 2007;1:69–74.
Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93.
Schweiger MR, Kerick M, Timmermann B, Albrecht MW, Borodina T, Parkhomchuk D, et al. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS One. 2009;4:e5548.
Acknowledgements
This study received financial support from the “Augustinus” and the “Anna og Dagny Hjerrilds” foundations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hansen, J., Lesnikova, I., Funder, A.M.D. et al. DNA and RNA analysis of blood and muscle from bodies with variable postmortem intervals. Forensic Sci Med Pathol 10, 322–328 (2014). https://doi.org/10.1007/s12024-014-9567-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12024-014-9567-2