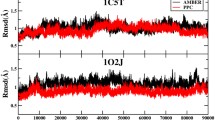
Abstract
MD simulation study of several peptides including a polyalanine, a helix (pdb:2I9M), and a leucine zipper were carried out to investigate hydrogen bond energetics using dynamic polarized protein-specific charge (DPPC) to account for the polarization effect in protein dynamics. Results show that the backbone hydrogen-bond strength is generally correlated with its specific local electrostatic environment, measured by the number of water molecules near the hydrogen bond in the first solvation shell. The correlation coefficient is found to be 0.89, 0.78, and 0.80, respectively, for polyalanine, 2I9M protein, and leucine zipper. In the polyalanine, the energies of the backbone hydrogen bonds are very similar to each other due to their similar local electrostatic environment. The current study helps demonstrate and support the understanding that hydrogen bonds are stronger in a hydrophobic surrounding than in a hydrophilic one. For comparison, the result from simulation using standard force field shows a much weaker correlation between hydrogen bond energy and local electrostatic environment due to the lack of polarization effect in the force field.
Similar content being viewed by others
References
Dill KA. Dominant forces in protein folding. Biochemistry, 1990, 29: 7133–7155
Honig B, Yang AS. Free energy balance in protein folding. Adv Prot Chem, 1995, 46: 27–58
Lumb KJ, Kim PS. A buried polar interaction imparts structural uniqueness in a designed heterodimeric coiled coil. Biochemistry, 1995, 34: 8642–8648
Myers JK, Pace CN. Hydrogen bonding stabilizes globular proteins. Biophys J, 1996, 71: 2033–2039
Petrey D, Honig B. Free energy determinants of tertiary structure and the evaluation of protein models. Protein Sci, 2000, 9: 2181–2191
Bolen DW, Rose GD. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu Rev Biochem, 2008, 77: 339–362
Mirsky AE, Pauling L. On the structure of native, denatured, and coagulated proteins. Proc Natl Acad Sci USA, 1936, 22: 439–447
Pauling L, Corey RB. Configurations of polypeptide chains with favored orientations around single bonds: two new pleated sheets. Proc Natl Acad Sci USA, 1951, 37: 729–740
Pauling L, Corey RB, Branson HR. The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci USA, 1951, 37: 205–211
Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Prot Chem, 1959, 14: 1–63
Klotz IM, Franzen JS. Hydrogen bonds between model peptide groups in solution. J Am Chem Soc, 1962, 84: 3461–3466
Epand RM, Scheraga HA. The influence of long-range interactions on the structure of myoglobin. Biochemistry, 1968, 7: 2864–2872
Taniuchi JH, Anfinsen CB. An experimental approach to the study of the folding of staphylococcal nuclease. J Biol Chem, 1969, 244: 3864–3875
Fersht AR. The hydrogen-bond in molecular recognition. Trends Biochem Sci, 1987, 12: 301–304
Baldwin RL. In search of the energetic role of peptide hydrogen bonds. J Biol Chem, 2003, 278: 17581–17588
Umeyama H, Morokuma K. The origin of hydrogen-bonding: an energy decomposition study. J Am Chem Soc, 1977, 99: 1316–1332
Fernández A, Berry RS. Extent of hydrogen-bond protection in folded proteints: a constraint on packing architectures. Biophys J, 2002, 83: 2475–2481
Némethy G, Steinberg IZ, Scheraga HA. Influence of water structure and of hydrophobic interactions on the strength of side-chain hydrogen bonds in proteins. Biopolymers, 1963, 1: 43–69
Gao J, Bosco DA, Powers ET, Kelly JW. Localized thermodynamic coupling between hydrogen bonding and microenvironment polarity significantly stabilizes proteins. Nat Struct Mol Biol, 2009, 16: 684–690
Ji CG, Mei Y, Zhang JZH. Developing polarized protein-specific charges for protein dynamics: MD free energy calculation of pKa shifts for Asp26/Asp20 in thioredoxin. Biophys J, 2008, 95: 1080–1088
Ji CG, Zhang JZH. Protein polarization is critical to stabilizing AF-2 and helix-2′ domains in ligand binding to PPAR-γ. J Am Chem Soc, 2008, 50: 17129–17133
Duan LL, Mei Y, Zhang QG, Zhang JZH. Intra-protein hydrogen bonding is dynamically stabilized by electronic polarization. J Chem Phys, 2009, 130: 115102
Tong Y, Ji CG, Mei Y, Zhang JZH. Simulation of NMR data reveals that proteins’ local structures are stabilized by electronic polarization. J Am Chem Soc, 2009, 131: 8636–8641
Duan LL, Mei Y, Zhang DW, Zhang QG, Zhang JZH. Folding of a helix at room temperature is critically aided by electrostatic polarization of intraprotein hydrogen bonds. J Am Chem Soc, 2010, 132: 11159–11164
Tong Y, Mei Y, Li YL, Ji CG, Zhang JZH. Electrostatic polarization interaction makes substantial contribution to free energy of avidin-biotin binding. J Am Chem Soc, 2010, 132: 5137–5142
Ji CG, Zhang JZH. Quantifying the stabilizing energy of the intraprotein hydrogen bond due to local mutation. J Phys Chem B, 2011, 115: 12230–12233
Ji CG, Zhang JZH. Understanding the molecular mechanism of enzyme dynamics of ribonuclease A through protonation/deprotonation of HIS48. J Am Chem Soc, 2011, 133: 17727–17737
Mei Y, Zhang DW, Duan LL, Zhang QG, Zhang JZH. Folding of EK peptide and its dependence on salt concentration and pH: a computational study. Sci China Chem, 2011, 54: 1974–1981
Uceda DP, Pastor MT, Salgado J, Lucena AP, Payá EP. Design of a bivalent peptide with two independent elements of secondary structure able to fold autonomously. J Pept Sci, 2008, 14: 845–854
Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. Embo J, 1994, 13: 2849–2861
Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA-binding proteins. Science, 1988, 240: 1759–1764
Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham III TE, DelBolt S, Ferguson D, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp Phys Commun, 1995, 91: 1–41
Pastor RW, Brooks BR, Szabo A. An analysis of the accuracy of langevin and molecular dynamics algorithm. Mol Phys, 1988, 65: 1409–1419
Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constrains: molecular dynamics of n-alkanes. J Comput Phys, 1977, 23: 327–341
Xiang Y, Duan LL, Zhang JZH. Protein’s electronic polarization contributes significantly to its catalytic function. J Chem Phys, 2011, 134: 205101
Li YL, Mei Y, Zhang DW, Xie DQ, Zhang JZH. Structure and dynamics of a dizinc metalloprotein: effect of charge transfer and polarization. J Phys Chem B, 2011, 115: 10154–10162
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Duan, L.L., Gao, Y., Ji, C.G. et al. Energetics of protein backbone hydrogen bonds and their local electrostatic environment. Sci. China Chem. 57, 1708–1715 (2014). https://doi.org/10.1007/s11426-014-5246-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-014-5246-0