Abstract
We argue that the overall behavior of tropomyosin on F-actin cannot be easily discerned by examining thin filaments reduced to their smallest interacting units. In isolation, the individual interactions of actin and tropomyosin, by themselves, are too weak to account for the specificity of the system. Instead the association of tropomyosin on actin can only be fully explained after considering the concerted action of the entire acto-tropomyosin system. We propose that the low K a describing tropomyosin:actin interaction, when taken together with the form-fitting complementarity of tropomyosin strands on F-actin and the tendency for tropomyosin to polymerize end-to-end, make possible unique thin filament functions both locally and at higher levels of filament organization.
Similar content being viewed by others
Introduction
Tropomyosin is a two-chained, coiled-coil protein that is found bound to actin filaments in most eukaryotic cells. The tropomyosin coiled-coil itself assumes a helical contour and thus represents a coiled coiled-coil. Tropomyosin molecules link end-to-end to form continuous strands that are wound tightly along the surface of helically disposed actin subunits of thin filaments (for reviews see Perry 2001; Brown and Cohen 2005; Hitchcock-DeGregori 2008; Lehman and Craig 2008). The presence of tropomyosin on actin strengthens thin filaments by increasing their rigidity and reducing their susceptibility to depolymerizing factors. Under the control of troponin and Ca2+ in skeletal and cardiac muscles, and perhaps other factors in most cellular systems including smooth muscles, tropomyosin participates in a molecular switching mechanism to regulate the traffic of myosin motors on actin filaments. Here access of myosin onto actin is gated by tropomyosin, with troponin in the case of striated muscle filaments playing the role of gatekeeper. This is accomplished by azimuthal movement of tropomyosin among different set positions along thin filaments, thereby either exposing or blocking myosin-binding sites on actin (Haselgrove 1972; Huxley 1972; Parry and Squire 1973; Lehman et al. 1994; Vibert et al. 1997; Poole et al. 2006). An intrinsic ability of tropomyosin molecules to undergo such shifts in position on actin at low energy cost is inherent to this steric mechanism (Lehman et al. 2000), and it is troponin that causes tropomyosin to favor one position or another depending on the concentration of Ca2+. This steric mechanism is not just limited to regulating myosin function. Thus, the presence of smooth muscle and non-muscle isoforms of tropomyosin on actin, possibly in concert with caldesmon (Hodgkinson et al. 1997; Hodgkinson 2000; Lehman et al. 2000), no doubt also gate access of a host of cellular actin-binding proteins onto thin filaments (Ishikawa et al. 1989, 1998; Blanchoin et al. 2001; Gunning et al. 2008). However, despite the crucial role played by tropomyosin in actin function, the properties of tropomyosin that contribute to its F-actin binding and lead to its regulatory movements on actin are incompletely understood. In this commentary, we propose a novel mechanism for the tropomyosin–actin association that we call “Gestaltbindung” or “Gestalt-binding” to contrast the interaction with more typically described lock-and-key models involving stereo-specific protein–protein binding. The use of the term is intended to connote several intertwined meanings. Gestalt, a German word for “form”, reflects our argument that the native three-dimensional form taken by tropomyosin is designed to match the actin filament surface, a complementarity which we regard as essential for actin-binding. As in Gestalt-theory [a psychological theory developed by von Ehrenfels (1890) and later by his student Wertheimer (1912)], we also argue that the behavior of the whole system cannot be inferred by examining single “atomistic” units (here single tropomyosin molecules) but only emerges from an understanding of the concerted action of the entire system, viz. the elongated tropomyosin strand as a whole on actin. Finally, we argue that mutations leading to perturbations in tropomyosin shape cause a deficit in “Gestaltbindung”, in which irrelevant conformers, lacking complementarity to the contours of actin, fail to bind to thin filaments, as if the system were divorced from its collective genomic memory.
Tropomyosin is a modular protein with seven pseudo-repeating units designed to bind to and match the polarity of seven adjacent actin monomers along a 385 Å path of the helical strands of actin filaments (Parry 1975; Stewart and McLachlan 1975; McLachlan and Stewart 1976; Phillips et al. 1986; Brown and Cohen 2005). The binding strength of individual tropomyosin molecules to F-actin is very weak [K a ~ 2–5 × 103 M−1, Wegner (1980)]. If each pseudo-repeat module binds actin with roughly equivalent affinity, then the K a of one repeat would be ~3, close to the binding strength of a single salt bridge in proteins, a value so low that thermal energy would readily cause dissociation. The low K a , atypical of actin-binding proteins, indicates that strong stereo-specific interaction is not a significant factor in F-actin–tropomyosin association.
Tropomyosin only binds to actin filaments because of the collective interactions conferred by linking tropomyosin end on end. Estimates for the equilibrium of the tropomyosin end-to-end association are modest [2–4.5 × 102, Wegner (1980)]. However, the overall effect of gluing tropomyosin molecules together over an F-actin substrate significantly raises the effective binding of tropomyosin to thin filaments by many orders of magnitude (in principle to K 25 a for 25 tropomyosin molecules linked in tandem over a 1 μm distance). Thus, it is hardly surprising that altered tropomyosin molecules that are incapable of forming end-to-end links cannot associate appreciably with F-actin (Johnson and Smillie 1977; Heald and Hitchcock-DeGregori 1988; Cho et al. 1990; Monteiro et al. 1994).
The modular design of tropomyosin
In addition to the continuously repeating, seven amino acid long “heptads” that define all α-helical coiled-coils, tropomyosin displays a unique longer-range period that is roughly forty amino acids long (Crick 1953; Stone and Smillie 1978; Parry 1975; Stewart and McLachlan 1975; McLachlan and Stewart 1976; Phillips et al. 1986). The latter accounts for the seven modular domains of tropomyosin, which traverse seven actin-binding partners along thin filaments. Other coiled-coils present in muscle that lack the 7-fold modular periodicity do not bind to actin (e.g. the rod portion of thick filament associated myosin and paramyosin in invertebrates). However, Nature, unlike the Pythagoreans, did not settle on seven-fold motifs as an ideal application of a perfect number. In fact, most somatic cells contain truncated tropomyosin isoforms with four to six ~40 amino acid-long actin-binding modules designed to bind to shorter stretches of thin filaments (Gunning et al. 2005, 2008).
A series of elegant experiments by the Hitchcock-DeGregori and Tobacman groups (Hitchcock-DeGregori and An 1996; Landis et al. 1999; Rosol et al. 2000; Hitchcock-DeGregori et al. 2001, 2002) showed that strict precision in the modular quantization of tropomyosin is essential for actin-binding. Tropomyosin was truncated by deleting one or more internal repeat modules. When the remaining modules were left intact to conform to the actin filament periodicity, these constructs still bound to actin. In contrast, when insertions or deletions were introduced that changed the lengths of one or another of the modules, the actin–tropomyosin alignment was corrupted and actin-binding diminished considerably. Similarly, substitution of random coils in place of coiled-coil modules abolished actin-binding.
Tropomyosin contains strategically placed acidic residues on the surface of each of its repeating modules. These no doubt interact electrostatically with positively charged residues present on the complementary flat face of actin subunits along thin filaments (Lorenz et al. 1995; Brown et al. 2005). Such an interaction is consistent with sensitivity of tropomyosin–actin binding to monovalent ion concentration (Eaton et al. 1975; Hill et al. 1992; Willadsen et al. 1992). Lacking a high-resolution crystal structure of the actin–tropomyosin complex, the positions of interfacial interactions cannot be plotted unambiguously. Nonetheless, in silico modeling provides strong support for the view that complementary surface charges are matched to each other (Lorenz et al. 1995; Brown et al. 2005; Brown and Cohen 2005).
Tropomyosin lies at a high radius on the actin filament
Lorenz, Holmes and colleagues built an atomic model of the actin–tropomyosin complex by a fitting a coiled coiled-coil structure that was based on the tropomyosin sequence against X-ray fibre diffraction patterns of oriented gels of F-actin–tropomyosin (Lorenz et al. 1995). The resulting model indicated that tropomyosin lies over the F-actin surface at a radius of ~39 Å (Fig. 1 A). In a subsequent study, fitting the atomic coordinates of tropomyosin to EM reconstructions of troponin–tropomyosin regulated thin filaments gave comparable values for the radius of tropomyosin [40 Å radius for Ca2+-treated filaments, 42 Å radius for Ca2+-free filaments (Poole et al. 2006)]. At an average radius of close to 40 Å from the center of the filament, tropomyosin is too far away to “touch” the actin surface and hence to interact strongly. At this distance, the closest distance between Cα atoms of actin and tropomyosin is ~10–11 Å. The distance separating actin and tropomyosin should preclude stereo-specific lock-and-key binding. In contrast, the separation is consistent with weak electrostatic interactions dominating, as the preponderance of evidence suggests. While notionally a pattern of repeating electrostatic [and possibly other weak (Brown and Cohen 2005)] interactions between tropomyosin and actin subunits is “specific” to the system, the attraction of single pseudo-repeat modules of tropomyosin and actin can hardly be considered much more specific than the localization of a few cations at a charged surface. Instead, we propose that the basis of the specificity depends crucially on the shape complementarity of tropomyosin and actin filaments.
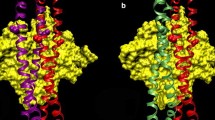
Structural contraints governing the binding of tropomyosin and F-actin. A Tropomyosin lies on F-actin at a high radius. Tropomyosin (red, yellow) modeled on F-actin (green, Holmes et al. 2003) as in Poole et al. (2006), shown as a (a) ribbon and (b) space-filling display. Note that, even at 39 Å radius, only long side chains of tropomyosin contact the surface of actin and that there is an extensive layer of water between tropomyosin and actin (also see Brown et al. 2005). The program PyMol (DeLano Scientific LLC) was used for the graphics display. B Fitting crystal structures of tropomyosin to the atomic model of tropomyosin. High-resolution crystal structures of tropomyosin (blue—the N-terminal fragment of tropomyosin (PDB ID#-1IC2), red—the mid-section of tropomyosin (PDB ID# 2b9c, Brown et al. 2001, 2005) were fitted to the Holmes/Lorenz coiled coiled-coil model of tropomyosin (green ribbon—α-carbon, Lorenz et al. 1995). Note that, with the exception of the ends of the crystal structures, which may have been unraveled, these the crystal structures fitted to the full length tropomyosin model very well. In turn, the tropomyosin model and the crystal structures also fit well to tropomyosin densities in low- and high-Ca2+ reconstructions of thin filaments (see panel C). C Fitting the tropomyosin coiled coiled-coil to thin filament reconstructions. The fitting of F-actin (blue, white, sky-blue) and the Holmes/Lorenz model of tropomyosin (red or yellow) into (a) high- and (b) low-Ca2+ reconstructions of thin filaments (transparent envelope) reconstituted from actin, tropomyosin and troponin. Note that, despite the movement of tropomyosin, the same unique tropomyosin model fitted equally well to both thin filament states. Figure from Poole et al. (2006) with permission
Shape complementarity of tropomyosin and actin filaments
As described, the binding site for tropomyosin on actin is 385 Å long and made of seven low affinity interactions. Surely the only reason that the tropomyosin binds to actin at all is because it assumes the right shape to interact with seven actins all at the same time. This shape can be deduced from high-resolution crystal structures of segments of the molecule that fit very well to contours of tropomyosin in the Lorenz–Holmes atomic model of actin–tropomyosin described above (Brown et al. 2001, 2005; Li et al. 2002, see Fig. 1 B.) These structures also dock extremely well within tropomyosin density envelopes in 3D reconstructions of both Ca2+-treated and Ca2+-free thin filaments (Poole et al. 2006; see Fig. 1 C). No obvious reconfiguration of tropomyosin is needed for the fittings. Thus independently determined tropomyosin structures display a conformation which is directly transposable onto the actin filament, without any apparent need for significant coiled coiled-coil conformational flexibility. Hence, these results indicate that the modular sequence complementary of tropomyosin and F-actin, described above, and the shape complementary, described here, are perfectly correlated. More specifically, these studies not only support the hypothesis that tropomyosin is designed to match the contours of actin but also that it is free to move over a flat actin interface without undergoing an obvious change in configuration (Poole et al. 2006).
The presence of roughly periodic groups of alanine clusters (Conway and Parry 1990) in tropomyosin causes local narrowing and bending of the coiled-coil protein (Brown et al. 2001; Brown and Cohen 2005, also see Nitanai et al. 2007; Minakata et al. 2008). This bending likely is partly responsible for the global coiled coiled-coil shape of tropomyosin, which fits so nicely to the F-actin surface. The B-factor associated with the alanine clusters in crystal structures is low, suggesting that the clusters and their surrounds, while bent, are relatively stable. Formally, alanine clusters can bend a fibrous protein in opposite orientations. In fact, there must be a bias in folding of the tropomyosin-coiled coiled-coil, since crystal structures of tropomyosin fit to right-handed helices that match the right-handed F-actin helix. Thus the whole molecule appears to bend collectively when forming the coiled coiled-coil.
We posit that the inherent helical contour of tropomyosin is an essential feature that facilitates the binding of the elongated protein on actin, weak as this interaction might be. Experimental support for this notion comes from studies showing that replacing core alanine residues, responsible for specific bending, with leucine and valine can reduce the binding of such tropomyosin variants to F-actin (Singh and Hitchcock-DeGregori 2003). This observation suggests that disrupting coiled-coil bending reconfigures the tropomyosin supercoil, which is followed by a concomitant loss of complementarity and hence Gestaltbindung to F-actin. Experimental alteration in tropomyosin pseudo-repeat lengths will also have the same effect. Whether or not such mutations cause changes in flexibility of tropomyosin (Singh and Hitchcock-DeGregori 2003, 2006) may not be as relevant a consideration.
End-to-end polymers of tropomyosin bind tightly to actin filaments
The head-to-tail polymerization of tropomyosin on the surface of actin filaments generates a larger scale level of thin filament organization that is functionally transformative. As mentioned, tropomyosin assembly and binding on actin is only effective once these end-to-end links are formed. In fact, experimental manipulation that interferes with tropomyosin strand formation (e.g. expressing unacetylated tropomyosin constructs or removing tropomyosin ends) fixes the system at the lower level of organization and prevents effective actin-binding (Johnson and Smillie 1977; Heald and Hitchcock-DeGregori 1988; Cho et al. 1990; Monteiro et al. 1994). There appear to be multiple mechanisms for ensuring that distinctive ends of different tropomyosin isoforms adhere to each other and several structural models have been offered for the process (McLachlan and Stewart 1976; Greenfield et al. 2006; Murakami et al. 2008). Understanding the specificity of the tropomyosin–tropomyosin connections is important. However, of possible greater importance is recognition that only minimum energy cost may be needed to bias tropomyosin into polymeric strands and thus allow the protein to work as part of an unbroken cooperative system, where Aristotle’s dictum, “The whole is more than the sum of its parts” applies. The Gestalt of the thin filament system as a whole can only then structure how the parts behave.
Clear and convincing evidence indicates that, in the complete absence of any accompanying actin-binding proteins, muscle and non-muscle isoforms of tropomyosin bind to actin with great precision and regularity (Lorenz et al. 1995; Lehman et al. 2000; Skoumpla et al. 2007; Maytum et al. 2008). Binding of the two proteins to each other is an innate property that is intrinsic to the design of tropomyosin and actin filaments. However, given the diversity of tropomyosin isoforms, particularly those in non-muscle cells (Gunning et al. 2005, 2008), it is not surprising that different tropomyosins show differences in their affinity for actin (Pittenger et al. 1995). This potentially could result from subtle changes in surface charge, in overall coiled coiled-coil shape and flexibility, or in the end-to-end molecular bonding of tropomyosin. It is also possible that extrinsic factors such as ancillary actin-binding proteins may modulate the conformational landscape of the actin filament to influence Gestalt-binding (cf. Hill et al. 1992). In addition, actin or tropomyosin binding proteins such as troponin-T and caldesmon may tether tropomyosin to actin, thus stabilizing assembled thin filaments (Hill et al. 1992; Pittenger et al. 1995).
Proposed scheme for tropomyosin binding to actin filaments
We envision that the following sequence takes place when tropomyosin assembles on actin filaments (see schematic, Fig. 2). Individual tropomyosin molecules transiently bind to F-actin filaments with an extremely low K a . The association is only possible because of the structural complementarity of the F-actin–tropomyosin interface, decreasing the entropic expense of the interaction. Lacking any definitive data, it is difficult to judge whether or not some limited amount of flexibility in either the tropomyosin supercoil or the F-actin filament facilitates or hampers this process. Since contours of the 40 nm long tropomyosin molecule represent a half turn of the actin helix; there is no need for extensive tropomyosin plasticity for the molecule to wrap around actin as is sometimes suggested. Thus, in principle, tropomyosin molecules can approach actin as fairly rigid coiled–coiled coils and be held roughly in place by electrostatics. Once a critical concentration of tropomyosin on actin is reached, tropomyosin will find partners (the next tropomyosin molecules along the filament) and link up to form polymers. It is only then that binding becomes appreciable (In solution, large entropic factors are likely to limit building even short tropomyosin oligomers. Quite the opposite should occur on the surface of actin where the entropic effects are more or less removed.). As tropomyosin polymers begin to populate the thin filaments, there will be a sorting process that favors the binding of long strings of end-to-end linked tropomyosin over shorter oligomers that are separated by gaps of tropomyosin-free actin. This stochastic mechanism will effectively zipper tropomyosin and actin together and hence lead to full decoration. While the binding of polymerized tropomyosin strands on fully decorated F-actin will be strong, the position of tropomyosin on actin at a local level will still be defined by weak electrostatic interactions and be easily perturbed by troponin, myosin and other proteins at low energy cost. Thus described, the Gestaltbindung of the thin filament system is well adapted to the physiological demands placed on it. No doubt, Gestaltbindung is a general feature that defines other biomolecular interactions as well, and it is hoped that the foregoing discussion will stimulate further structural experimentation and in silico modeling of such systems.
Schematic representation of tropomyosin binding to F-actin. a tropomyosin molecules (multicolored curved rods) in initial mixtures of F-actin (double chain of beads) and tropomyosin are largely monomeric, b tropomyosin binds actin with very low K a , and only because it has the correct shape to interact with seven successive actin monomers all at the same time, c tropomyosin molecules form end-to-end bonds with neighboring tropomyosin molecules on F-actin, d once a critical concentration is reached, tropomyosin forms polymers and a sorting process occurs between long and short tropomyosin strands leading to full saturation of tropomyosin on F-actin (e)
References
Blanchoin L, Pollard TD, Hitchcock-DeGregori SE (2001) Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol 11:1300–1304. doi:10.1016/S0960-9822(01)00395-5
Brown JH, Cohen C (2005) Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem 71:121–159. doi:10.1016/S0065-3233(04)71004-9
Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C (2001) Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci USA 98:8496–8501. doi:10.1073/pnas.131219198
Brown JH, Zhou Z, Reshetnikova L, Robinson H, Yammani RD, Tobacman LS, Cohen C (2005) Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci USA 102:18878–18883. doi:10.1073/pnas.0509269102
Cho YJ, Liu J, Hitchcock-DeGregori SE (1990) The amino terminus of muscle tropomyosin is a major determinant for function. J Biol Chem 265:538–545
Conway JF, Parry DA (1990) Structural features in the heptad substructure and longer range repeats of two-stranded alpha-fibrous proteins. Int J Biol Macromol 12:328–334. doi:10.1016/0141-8130(90)90023-4
Crick FHC (1953) The packing of alpha-helices: simple coiled-coils. Acta Crystallogr 6:689–697. doi:10.1107/S0365110X53001964
Eaton BL, Kominz DR, Eisenberg E (1975) Correlation between the inhibition of the acto-heavy meromyosin ATPase and the binding of tropomyosin to F-actin: effects of Mg2+, KCl, troponin I, and troponin C. Biochemistry 14:2718–2725. doi:10.1021/bi00683a025
Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE (2006) Solution NMR structure of the junction between tropomyosin molecules: implications for actin-binding and regulation. J Mol Biol 364:80–96. doi:10.1016/j.jmb.2006.08.033
Gunning PW, Schevzov G, Kee AJ, Hardeman EC (2005) Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol 15:333–341. doi:10.1016/j.tcb.2005.04.007
Gunning PW, O’Neill G, Hardemann EC (2008) Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev 88:1–35. doi:10.1152/physrev.00001.2007
Haselgrove JC (1972) X-ray evidence for a conformational change in actin-containing filaments of vertebrate striated muscle. Cold Spring Harb Symp Quant Biol 37:341–352
Heald RW, Hitchcock-DeGregori SE (1988) The structure of the amino terminus of tropomyosin is critical for binding to actin in the absence and presence of troponin. J Biol Chem 263:5254–5259
Hill LE, Mehegan JP, Butters CA, Tobacman LS (1992) Analysis of troponin–tropomyosin binding to actin. Troponin does not promote interactions between tropomyosin molecules. J Biol Chem 267:16106–16113
Hitchcock-DeGregori SE (2008) Tropomyosin: function follows form. Tropomyosin and the steric mechanism of muscle regulation. In: Gunning P (ed) Tropomyosin. Landes BioScience, Austin, TX, Adv Exp Med Biol 644:60–72
Hitchcock-DeGregori SE, An Y (1996) Integral repeats and a continuous coiled coil are required for binding of striated muscle tropomyosin to the regulated actin filament. J Biol Chem 271:3600–3603. doi:10.1074/jbc.271.8.4236
Hitchcock-DeGregori SE, Song Y, Moraczewska J (2001) Importance of internal regions and the overall length of tropomyosin for actin-binding and regulatory function. Biochemistry 40:2104–2112. doi:10.1021/bi002421z
Hitchcock-DeGregori SE, Song Y, Greenfield NJ (2002) Functions of tropomyosin’s periodic repeats. Biochemistry 41:15036–15044. doi:10.1021/bi026519k
Hodgkinson JL (2000) Actin and smooth muscle regulatory proteins: a structural perspective. J Muscle Res Cell Motil 32:115–130. doi:10.1023/A:1005697301043
Hodgkinson JL, Marston SB, Craig R, Vibert P, Lehman W (1997) Three-dimensional image reconstruction of reconstituted smooth muscle thin filaments: Effects of caldesmon. Biophys J 72:2398–2404
Holmes KC, Angert I, Kull FJ, Jahn W, Schröder RR (2003) Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425:423–427. doi:10.1038/nature02005
Huxley HE (1972) Structural changes in actin-and myosin-containing filaments during contraction. Cold Spring Harb Symp Quant Biol 37:361–376
Ishikawa R, Yamashiro S, Matsumura F (1989) Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem 264:7490–7497
Ishikawa R, Yamashiro S, Kohama K, Matsumura F (1998) Regulation and actin-binding activities of fascin by caldesmon coupled with tropomyosin. J Biol Chem 273:26991–26997. doi:10.1074/jbc.273.41.26991
Johnson P, Smillie LB (1977) Polymerizability of rabbit skeletal tropomyosin: effects of enzymic and chemical modifications. Biochemistry 16:2264–2269. doi:10.1021/bi00629a035
Landis C, Back N, Homsher E, Tobacman LS (1999) Effects of tropomyosin internal deletions on thin filament function. J Biol Chem 274:31279–31285. doi:10.1074/jbc.274.44.31279
Lehman W, Craig R (2008) Tropomyosin and the steric mechanism of muscle regulation. In: Gunning P (ed) Tropomyosin. Landes BioScience, Austin, TX, Adv Exp Med Biol 644:95–109
Lehman W, Craig R, Vibert P (1994) Ca2+-induced tropomyosin movement in Limulus thin filaments revealed by three- dimensional reconstruction. Nature 368:65–67. doi:10.1038/368065a0
Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, Craig R (2000) Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J Mol Biol 302:593–606. doi:10.1006/jmbi.2000.4080
Li Y, Mui S, Brown JH, Strand J, Reshetnikova L, Tobacman LS, Cohen C (2002) The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc Natl Acad Sci USA 99:7378–7383. doi:10.1073/pnas.102179999
Lorenz M, Poole KJV, Popp D, Rosenbaum G, Holmes KC (1995) An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin–tropomyosin gels. J Mol Biol 246:108–119. doi:10.1006/jmbi.1994.0070
Maytum R, Hatch V, Konrad M, Lehman W, Geeves MA (2008) Ultra short yeast tropomyosins show novel myosin regulation. J Biol Chem 283:1902–1910. doi:10.1074/jbc.M708593200
McLachlan AD, Stewart M (1976) The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J Mol Biol 103:271–298. doi:10.1016/0022-2836(76)90313-2
Minakata S, Maeda K, Oda N, Wakabayashi K, Nitanai Y, Maéda Y (2008) Two-crystal structures of tropomyosin C-terminal fragment 176–273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophys J 95:710–719. doi:10.1529/biophysj.107.126144
Monteiro PB, Lataro RC, Ferro JA, Reinach FC (1994) Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J Biol Chem 269:10461–10466
Murakami K, Stewart M, Nozawa K, Tomii K, Kudou N, Igarashi N, Shirakihara Y, Wakatsuki S, Yasunaga T, Wakabayashi T (2008) Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T. Proc Natl Acad Sci USA 105:7200–7205. doi:10.1073/pnas.0801950105
Nitanai Y, Minakata S, Maéda K, Oda N, Maéda Y (2007) Crystal structures of tropomyosin: flexible coiled-coil. Adv Exp Med Biol 592:137–151. doi:10.1007/978-4-431-38453-3_13
Parry DA (1975) Analysis of the primary sequence of alpha-tropomyosin from rabbit skeletal muscle. J Mol Biol 98:519–535. doi:10.1016/S0022-2836(75)80084-2
Parry DAD, Squire JM (1973) Structural role of tropomyosin in muscle regulation, analysis of X-ray patterns from relaxed and contracting muscles. J Mol Biol 75:33–55. doi:10.1016/0022-2836(73)90527-5
Perry SV (2001) What is the role of tropomyosin in the regulation of muscle contraction? J Muscle Res Cell Motil 24:593–596
Phillips JGN, Fillers JP, Cohen C (1986) Tropomyosin crystal structure and muscle regulation. J Mol Biol 192:111–131. doi:10.1016/0022-2836(86)90468-7
Pittenger MF, Kistler A, Helfman DM (1995) Alternatively spliced exons of the β tropomyosin gene exhibit different affinities and effects with nonmuscle caldesmon. J Cell Sci 108:3253–3265
Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Tobacman LS, Lehman W, Holmes KC (2006) A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol 155:273–284. doi:10.1016/j.jsb.2006.02.020
Rosol M, Lehman W, Landis C, Butters C, Craig R, Tobacman LS (2000) Three-dimensional reconstruction of thin filaments containing mutant tropomyosin. Biophys J 78:918–926
Singh A, Hitchcock-DeGregori SE (2003) Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin-binding. Biochemistry 42:14114–14121. doi:10.1021/bi0348462
Singh A, Hitchcock-DeGregori SE (2006) Dual requirement for flexibility and specificity for binding of the coiled-coil tropomyosin to its target, actin. Structure 14:43–50. doi:10.1016/j.str.2005.09.016
Skoumpla K, Coulton AT, Lehman W, Geeves MA, Mulvihill DP (2007) Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J Cell Sci 120:1635–1645. doi:10.1242/jcs.001115
Stewart M, McLachlan AD (1975) Fourteen actin-binding sites on tropomyosin? Nature 257:331–333. doi:10.1038/257331a0
Stone D, Smillie LB (1978) The amino acid sequence of rabbit skeletal alpha-tropomyosin. The NH2-terminal half and complete sequence. J Biol Chem 253:1137–1148
Vibert P, Craig R, Lehman W (1997) Steric-model for activation of muscle thin filaments. J Mol Biol 266:8–14. doi:10.1006/jmbi.1996.0800
von Ehrenfels CF (1890) Über Gestaltqualitäten. Vierteljahrsschrift für wissenschaftliche Philosophie 14:249–292
Wegner A (1980) The interaction of alpha, alpha-and alpha, beta-tropomyosin with actin filaments. FEBS Lett 119:245–248. doi:10.1016/0014-5793(80)80263-8
Wertheimer M (1912) Experimentelle Studien über das Sehen von Bewegung. Z Psychol Z Angew Psychol 61:161–265
Willadsen KA, Butters CA, Hill LE, Tobacman LS (1992) Effects of the amino-terminal regions of tropomyosin and troponin T on thin filament assembly. J Biol Chem 267:23746–23752
Acknowledgments
We thank Ms. Karen Moore (Moore Design) for artwork in Fig. 2. We also thank the restaurant staff at Schloss Schwetzingen for allowing the authors to first discuss Gestaltbindung a bit past closing hours. This research was supported by grants from the National Institutes of Health to W.L. (HL36153, HL86655).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Holmes, K.C., Lehman, W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil 29, 213–219 (2008). https://doi.org/10.1007/s10974-008-9157-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10974-008-9157-6