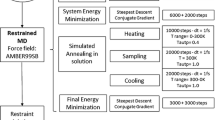
Abstract
A method is introduced to represent an ensemble of conformers of a protein by a single structure in torsion angle space that lies closest to the averaged Cartesian coordinates while maintaining perfect covalent geometry and on average equal steric quality and an equally good fit to the experimental (e.g. NMR) data as the individual conformers of the ensemble. The single representative ‘regmean structure’ is obtained by simulated annealing in torsion angle space with the program CYANA using as input data the experimental restraints, restraints for the atom positions relative to the average Cartesian coordinates, and restraints for the torsion angles relative to the corresponding principal cluster average values of the ensemble. The method was applied to 11 proteins for which NMR structure ensembles are available, and compared to alternative, commonly used simple approaches for selecting a single representative structure, e.g. the structure from the ensemble that best fulfills the experimental and steric restraints, or the structure from the ensemble that has the lowest RMSD value to the average Cartesian coordinates. In all cases our method found a structure in torsion angle space that is significantly closer to the mean coordinates than the alternatives while maintaining the same quality as individual conformers. The method is thus suitable to generate representative single structure representations of protein structure ensembles in torsion angle space. Since in the case of NMR structure calculations with CYANA the single structure is calculated in the same way as the individual conformers except that weak positional and torsion angle restraints are added, we propose to represent new NMR structures by a ‘regmean bundle’ consisting of the single representative structure as the first conformer and all but one original individual conformers (the original conformer with the highest target function value is discarded in order to keep the number of conformers in the bundle constant). In this way, analyses that require a single structure can be carried out in the most meaningful way using the first model, while at the same time the additional information contained in the ensemble remains available.






Similar content being viewed by others
References
Antuch W, Güntert P, Wüthrich K (1996) Ancestral βγ-crystallin precursor structure in a yeast killer toxin. Nat Struct Biol 3:662–665
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Betancourt MR, Skolnick J (2001) Finding the needle in a haystack: educing native folds from ambiguous ab initio protein structure predictions. J Comput Chem 22:339–353
Bianchetti CM, Blouin GC, Bitto E, Olson JS, Phillips GN (2010) The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At 1g79260.1. Proteins 78:917–931
Bowie JU, Lüthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known 3-dimensional structure. Science 253:164–170
Calzolai L, Lysek DA, Perez DR, Güntert P, Wüthrich K (2005) Prion protein NMR structures of chickens, turtles, and frogs. Proc Natl Acad Sci USA 102:651–655
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66:12–21
Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32:W615–W619
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383
Dukka BKC (2009) Improving consensus structure by eliminating averaging artifacts. BMC Struct Biol 9:12
Furnham N, Blundell TL, DePristo MA, Terwilliger TC (2006) Is one solution good enough? Nat Struct Mol Biol 13:184–185
Güntert P (2003) Automated NMR protein structure calculation. Prog Nucl Magn Reson Spectrosc 43:105–125
Güntert P (2009) Automated structure determination from NMR spectra. Eur Biophys J 38:129–143
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298
Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227
Hooft RWW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381:272
Horst R, Damberger F, Luginbühl P, Güntert P, Peng G, Nikonova L, Leal WS, Wüthrich K (2001) NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci USA 98:14374–14379
Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Ono AM, Güntert P (2006) Optimal isotope labelling for NMR protein structure determinations. Nature 440:52–57
Kelley LA, Gardner SP, Sutcliffe MJ (1996) An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng 9:1063–1065
Kirchner DK, Güntert P (2011) Objective identification of residue ranges for the superposition of protein structures. BMC Bioinform 12:170
Kolbe M, Besir H, Essen LO, Oesterhelt D (2000) Structure of the light-driven chloride pump halorhodopsin at 1.8 Å resolution. Science 288:1390–1396
Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55
Koradi R, Billeter M, Güntert P (2000) Point-centered domain decomposition for parallel molecular dynamics simulation. Comput Phys Commun 124:139–147
Kurpiewska K, Font J, Ribó M, Vilanova M, Lewiński K (2009) X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability. Proteins 77:658–669
Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486
Linge JP, Williams MA, Spronk CAEM, Bonvin AMJJ, Nilges M (2003) Refinement of protein structures in explicit solvent. Proteins 50:496–506
López-Méndez B, Güntert P (2006) Automated protein structure determination from NMR spectra. J Am Chem Soc 128:13112–13122
López-Méndez B, Pantoja-Uceda D, Tomizawa T, Koshiba S, Kigawa T, Shirouzu M, Terada T, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Hirota H, Yoshida M, Tanaka A, Osanai T, Seki M, Shinozaki K, Yokoyama S, Güntert P (2004) NMR assignment of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana. J Biomol NMR 29:205–206
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with 3-dimensional profiles. Nature 356:83–85
Luginbühl P, Güntert P, Billeter M, Wüthrich K (1996) The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J Biomol NMR 8:136–146
Morris AL, Macarthur MW, Hutchinson EG, Thornton JM (1992) Stereochemical quality of protein structure coordinates. Proteins 12:345–364
Nilges M, Clore GM, Gronenborn AM (1988) Determination of three-dimensional structures of proteins from interproton distance data by hybrid distance geometry-dynamical simulated annealing calculations. FEBS Lett 229:317–324
Ohnishi S, Güntert P, Koshiba S, Tomizawa T, Akasaka R, Tochio N, Sato M, Inoue M, Harada T, Watanabe S, Tanaka A, Shirouzu M, Kigawa T, Yokoyama S (2007) Solution structure of an atypical WW domain in a novel β-clam-like dimeric form. FEBS Lett 581:462–468
Pääkkönen K, Tossavainen H, Permi P, Rakkolainen H, Rauvala H, Raulo E, Kilpeläinen I, Güntert P (2006) Solution structures of the first and fourth TSR domains of F-spondin. Proteins 64:665–672
Pantoja-Uceda D, López-Méndez B, Koshiba S, Kigawa T, Shirouzu M, Terada T, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Hirota H, Yoshida M, Tanaka A, Osanai T, Seki M, Shinozaki K, Yokoyama S, Güntert P (2004) NMR assignment of the hypothetical rhodanese domain At4g01050 from Arabidopsis thaliana. J Biomol NMR 29:207–208
Pantoja-Uceda D, López-Méndez B, Koshiba S, Inoue M, Kigawa T, Terada T, Shirouzu M, Tanaka A, Seki M, Shinozaki K, Yokoyama S, Güntert P (2005) Solution structure of the rhodanese homology domain At4g01050(175–295) from Arabidopsis thaliana. Protein Sci 14:224–230
Pellecchia M, Sem DS, Wüthrich K (2002) NMR in drug discovery. Nat Rev Drug Discov 1:211–219
Ponder JW, Case DA (2003) Force fields for protein simulations. Adv Prot Chem 66:27–85
Reckel S, Gottstein D, Stehle J, Löhr F, Verhoefen MK, Takeda M, Silvers R, Kainosho M, Glaubitz C, Wachtveitl J, Bernhard F, Schwalbe H, Güntert P, Dötsch V (2011) Solution NMR structure of proteorhodopsin. Angew Chem 50:11942–11946
Rosato A, Aramini J, Arrowsmith C, Bagaria A, Baker D, Cavalli A, Doreleijers JF, Eletsky A, Giachetti A, Guerry P, Gutmanas A, Güntert P, F. HY, Herrmann T, Huang YJ, Jaravine V, Jonker HRA, Kennedy MA, Lange OF, Liu G, Malliavin TE, Mani R, Mao B, Montelione GT, Nilges M, Rossi P, van der Schot G, Schwalbe H, Szyperski T, Vendruscolo M, Vernon R, Vranken WF, de Vries S, Vuister GW, Wu B, Yang Y, Bonvin AMJJ (2012) Blind testing of routine, fully automated determination of protein structures from NMR data. Structure 8:227–236
Rosato A, Bagaria A, Baker D, Bardiaux B, Cavalli A, Doreleijers JF, Giachetti A, Guerry P, Güntert P, Herrmann T, Huang YJ, Jonker HRA, Mao B, Malliavin TE, Montelione GT, Nilges M, Raman S, van der Schot G, Vranken WF, Vuister GW, Bonvin AMJJ (2009) CASD-NMR: critical assessment of automated structure determination by NMR. Nat Methods 6:625–626
Rotkiewicz P, Skolnick J (2008) Fast procedure for reconstruction of full-atom protein models from reduced representations. J Comput Chem 29:1460–1465
Schwieters CD, Clore GM (2002) Reweighted atomic densities to represent ensembles of NMR structures. J Biomol NMR 23:221–225
Scott A, Pantoja-Uceda D, Koshiba S, Inoue M, Kigawa T, Terada T, Shirouzu M, Tanaka A, Sugano S, Yokoyama S, Güntert P (2004) NMR assignment of the SH2 domain from the human feline sarcoma oncogene FES. J Biomol NMR 30:463–464
Scott A, Pantoja-Uceda D, Koshiba S, Inoue M, Kigawa T, Terada T, Shirouzu M, Tanaka A, Sugano S, Yokoyama S, Güntert P (2005) Solution structure of the Src homology 2 domain from the human feline sarcoma oncogene Fes. J Biomol NMR 31:357–361
Sippl MJ (1993) Recognition of errors in 3-dimensional structures of proteins. Proteins 17:355–362
Sutcliffe MJ (1993) Representing an ensemble of NMR-derived protein structures by a single structure. Protein Sci 2:936–944
Thomas D, Pastore A (2005) WHEATSHEAF: an algorithm to average protein structure ensembles. Acta Crystallogr D 61:112–116
Wallner B, Elofsson A (2003) Can correct protein models be identified? Protein Sci 12:1073–1086
Wimmer R, Herrmann T, Solioz M, Wüthrich K (1999) NMR structure and metal interactions of the CopZ copper chaperone. J Biol Chem 274:22597–22603
Zhang Y, Skolnick J (2004) SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem 25:865–871
Zhao DQ, Jardetzky O (1994) An assessment of the precision and accuracy of protein structures determined by NMR: dependence on distance errors. J Mol Biol 239:601–607
Acknowledgments
We gratefully acknowledge financial support by the Lichtenberg program of the Volkswagen Foundation and by a Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Bimodal averages and standard deviation of torsion angles
If the values \( S = \{ \phi_{1} , \ldots ,\phi_{n} \} \) of a torsion angle ϕ are clustered in two separate regions it makes little sense to determine an average value. Instead it is meaningful to split the set S into two disjoint subsets S 1 and S 2 for the purpose of computing two bimodal average values \( {{\overline{\phi}}_{1}} = \arg \sum\nolimits_{{k \in s_{1} }} {e^{{i\phi_{k} }} } \) and \( {{\overline{\phi}}_{1}} = \arg \sum\nolimits_{{k \in s_{2} }} {e^{{i\phi_{k} }} } \) of the torsion angle values in S 1 and S 2, respectively. The choice of S 1 and S 2 is optimal if it minimizes the bimodal standard deviation
that results from summing for each torsion angle value ϕ k the squared deviation from the closer of the two bimodal average values \( {{\overline{\phi}}_{1}} \) and \( {{\overline{\phi}}_{2}} \), taking into account the periodicity.
It would be computationally inefficient to evaluate \( \sigma_{\phi }^{(2)} \) for each of the 2n possible choices of the subsets S 1 and S 2. To determine a good approximation of the optimal bimodal average values in polynomial time, we first calculate the n × n matrix of torsion angle differences \( \Updelta \phi_{ij} = \min \left( {\left| {\phi_{i} - \phi_{j} } \right|,2\pi - \left| {\phi_{i} - \phi_{j} } \right|} \right) \). For all pairs (i, j) with \( \Updelta \phi_{ij} > \pi /4 \) (to avoid splitting into two hardly separated clusters), we compute \( \widetilde{{\phi_{1} }} = \arg \sum\nolimits_{{k:\Updelta \phi_{ki} \le \Updelta \phi_{kj} }} {e^{{i\phi_{k} }} } \) and \( \widetilde{{\phi_{2} }} = \arg \sum\nolimits_{{k:\Updelta \phi_{ki} > \Updelta \phi_{kj} }} {e^{{i\phi_{k} }} } \). (In the exponential functions i denotes the imaginary unit \( \sqrt {-1,} \) otherwise the index i.) The deviations of the individual torsion angle values \( \phi_{k} \) from \( \widetilde{{\phi_{1} }} \) and \( \widetilde{{\phi_{2} }} \) are given by \( \delta_{1k} = \min \left( {\left| {\phi_{k} - \widetilde{{\phi_{1} }}} \right|,2\pi - \left| {\phi_{k} - \widetilde{{\phi_{1} }}} \right|} \right) \) and \( \delta_{2k} = \min \left( {\left| {\phi_{k} - \widetilde{{\phi_{2} }}} \right|,2\pi - \left| {\phi_{k} - \widetilde{{\phi_{2} }}} \right|} \right) \) for k = 1, …, n. The corresponding subsets are \( S_{1} = \{ k\left| {\delta_{1k} \le \delta_{2k} } \right.\} \) and \( S_{2} = \{ k\left| {\delta_{1k} > \delta_{2k} } \right.\} \). We choose the optimal subsets S 1 and S 2 from the pair (i, j) that yields the largest value of \( \left| {\sum\nolimits_{{k \in S_{1} }} {e^{{i\phi_{k} }} } } \right| + \left| {\sum\nolimits_{{k \in S_{2} }} {e^{{i\phi_{k} }} } } \right| \) to obtain the bimodal average values \( {{\overline{\phi}}_{1}} \) and \( {{\overline{\phi}}_{2}} \). If S 2 contains more elements than S 1, we exchange the values of \( {{\overline{\phi}}_{1}} \) and \( {{\overline{\phi}}_{2}} \) such that \( {{\overline{\phi}}_{1}} \) always corresponds to the cluster with the larger number of elements.
Rights and permissions
About this article
Cite this article
Gottstein, D., Kirchner, D.K. & Güntert, P. Simultaneous single-structure and bundle representation of protein NMR structures in torsion angle space. J Biomol NMR 52, 351–364 (2012). https://doi.org/10.1007/s10858-012-9615-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-012-9615-8