Abstract
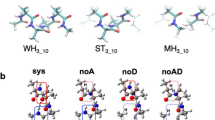
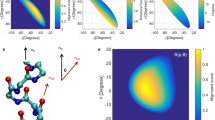
We performed density functional calculations of backbone 15N chemical shielding tensors in selected helical residues of protein G. Here we describe a computationally efficient methodology to include most of the important effects in the calculation of chemical shieldings of backbone 15N. We analyzed the role of long-range intra-protein electrostatic interactions by comparing models with different complexity in vacuum and in charge field. Our results show that the dipole moment of the α-helix can cause significant deshielding of 15N; therefore, it needs to be considered when calculating 15N chemical shielding. We found that it is important to include interactions with the side chains that are close in space when the charged form for ionizable side chains is adopted in the calculation. We also illustrate how the ionization state of these side chains can affect the chemical shielding tensor elements. Chemical shielding calculations using a 8-residue fragment model in vacuum and adopting the charged form of ionizable side chains yield a generally good agreement with experimental data.





Similar content being viewed by others
References
Bader R (2009) Utilizing the charge field effect on amide N-15 chemical shifts for protein structure validation. J Phys Chem B 113:347–358
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A 38:3098–3100
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Cai L, Fushman D, Kosov DS (2008) Density functional calculations of N-15 chemical shifts in solvated dipeptides. J Biomol NMR 41:77–88
Chesnut DB, Rusiloski BE, Moore KD, Egolf DA (1993) Use of locally dense basis-sets for nuclear magnetic resonance shielding calculations. J Comput Chem 14:1364–1375
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force-field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117:5179–5197
Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302
de Dios AC, Oldfield E (1993) Methods for computing nuclear magnetic resonance chemical shielding in large systems—multiple cluster and charge field approaches. Chem Phys Lett 205:108–116
de Dios AC, Pearson JG, Oldfield E (1993a) Chemical shifts in proteins—an ab initio study of C-13 nuclear magnetic resonance chemical shielding in glycine, alanine, and valine residues. J Am Chem Soc 115:9768–9773
de Dios AC, Pearson JG, Oldfield E (1993b) Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science 260:1491–1496
de Dios AC, Laws DD, Oldfield E (1994) Predicting C-13 nuclear magnetic resonance chemical shielding tensors in zwitterionic L-threonine and L-tyrosine via quantum chemistry. J Am Chem Soc 116:7784–7786
Ditchfield R (1974) Self-consistent perturbation theory of diamagnetism. 1. Gauge-invariant LCAO method for NMR chemical shifts. Mol Phys 27:789–807
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian 03, revision C.02. Gaussian Inc., Wallingford
Fushman D, Cowburn D (2001) Nuclear magnetic resonance relaxation in determination of residue-specific N-15 chemical shift tensors in proteins in solution: protein dynamics, structure, and applications of transverse relaxation optimized spectroscopy. Methods Enzymol 339:109–126
Fushman D, Tjandra N, Cowburn D (1998) Direct measurement of N-15 chemical shift anisotropy in solution. J Am Chem Soc 120:10947–10952
Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM (1991) A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 253:657–661
Hall JB, Fushman D (2006) Variability of the 15N chemical shielding tensors in the B3 domain of protein G from 15N relaxation measurements at several fields. Implications for backbone order parameters. J Am Chem Soc 128:7855–7870
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Le HB, Oldfield E (1996) Ab initio studies of amide-N-15 chemical shifts in dipeptides: applications to protein NMR spectroscopy. J Phys Chem 100:16423–16428
Lee CT, Yang WT, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron-density. Phys Rev B 37:785–789
Lipsitz RS, Tjandra N (2003) 15N chemical shift anisotropy in protein structure refinement and comparison with NH residual dipolar couplings. J Magn Reson 164:171–176
Nadaud PS, Helmus JJ, Jaroniec CP (2007) C-13 and N-15 chemical shift assignments and secondary structure of the B3 immunoglobulin-binding domain of streptococcal protein G by magic-angle spinning solid-state NMR spectroscopy. Biomol NMR Assign 1:117–120
Shen Y, Bax A (2007) Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR 38:289–302
Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA 105:4685–4690
Shen Y, Vernon R, Baker D, Bax A (2009) De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR 43:63–78
Ulmer TS, Ramirez BE, Delaglio F, Bax A (2003) Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy. J Am Chem Soc 125:9179–9191
Vila JA, Scheraga HA (2007) Factors affecting the use of 13C(alpha) chemical shifts to determine, refine, and validate protein structures. Proteins 71:641–654
Wolinski K, Hinton JF, Pulay P (1990) Efficient implementation of the Gauge-independent toamic orbital method for NMR chemical-shift calculations. J Am Chem Soc 112:8251–8260
Wylie BJ, Sperling LJ, Frericks HL, Shah GJ, Franks WT, Rienstra CM (2007) Chemical-shift anisotropy measurements of amide and carbonyl resonances in a microcrystalline protein with slow magic-angle spinning NMR spectroscopy. J Am Chem Soc 129:5318–5319
Wylie BJ, Schwieters CD, Oldfield E, Rienstra CM (2009) Protein structure refinement using 13C alpha chemical shift tensors. J Am Chem Soc 131:985–992
Xu XP, Case DA (2001) Automated prediction of N-15, C-13(alpha), C-13(beta) and C-13 chemical shifts in proteins using a density functional database. J Biomol NMR 21:321–333
Xu XP, Case DA (2002) Probing multiple effects on 15N, 13C alpha, 13C beta, and 13C chemical shifts in peptides using density functional theory. Biopolymers 65:408–423
Zuiderweg ERP (2002) Mapping protein–protein interactions in solution by NMR Spectroscopy. Biochemistry 41:1–7
Acknowledgments
Supported by NIH grant GM065334 to DF and by American Chemical Society Petroleum Research Fund (44481-G6) and Francqui Foundation to DSK. We thank Gabriel Cornilescu, Chad Rienstra, and Ben Wylie for making available detailed experimental chemical shift data for comparison.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, L., Fushman, D. & Kosov, D.S. Density functional calculations of chemical shielding of backbone 15N in helical residues of protein G. J Biomol NMR 45, 245–253 (2009). https://doi.org/10.1007/s10858-009-9358-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9358-3