Abstract
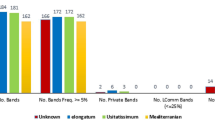
Little is known about the genetic diversity of pale flax (Linum bienne Mill.), the wild progenitor of cultivated flax (L. usitatissimum L.), and ex situ germplasm of pale flax was scarce. Effort was made to collect 34 pale flax accessions and five landrace accessions of cultivated flax in Turkey. The inter simple sequence repeat (ISSR) technique was applied to characterize this set of flax germplasm, along with one Turkish cultivar, one Russian cultivar, five winter and four dehiscent type accessions of cultivated flax. Twenty-four ISSR primer pairs detected a total of 311 DNA fragments, of which 298 bands were polymorphic across 493 flax samples (roughly 10 samples per accession). These polymorphic bands had frequencies ranging from 0.002 to 0.998 and averaging 0.38. Accession-specific ISSR variation (Fst values) ranged from 0.469 to 0.514 and averaged 0.493. There was 49.3% ISSR variation resided among these 50 accessions, 35.9% harbored among landrace, winter, dehiscent types of cultivated flax and pale flax, and 38.2% present among 34 pale flax accessions. Pale flax displayed more ISSR variation than landraces and dehiscent type, but less than winter type, of cultivated flax. Clustering 493 individual plants revealed that these assayed plants were largely grouped according to their plant types and that pale flax was genetically more close to the dehiscent type, followed by the winter type and landrace, of cultivated flax. Pale flax collected within the geographic range of 180 km displayed a significant spatial genetic autocorrelation. Genetic distances among the pale flax accessions were significantly associated with their geographic distances and elevation differences. These findings are significant for understanding flax domestication and its primary gene pool.




Similar content being viewed by others
References
Allaby RG, Peterson GW, Merriwether A, Fu YB (2005) Evidence of the domestication history of flax (Linum usitatissimum) from genetic diversity of the sad2 locus. Theor Appl Genet 112:58–65
Bhagyawant SS, Srivastava N (2008) Genetic fingerprinting of chickpea (Cicer arietinum L.) germplasm using ISSR markers and their relationships. Afr J Biotechnol 7:4428–4431
Campbell CD, Procunier JD, Oomah BD, Kenaschuk EO, Dribnenki JCP, Rashid KY (1995) Analysis of genetic relationships in flax (Linum usitatissimum) using RAPD markers. In: Proceedings of 56th Flax Institute of the United States, pp 177–181, Fargo, ND, USA
Cloutier S, Niu X, Datla R, Duguid S (2009) Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet 119:53–63
Diederichsen A (2007) Ex situ collection of cultivated flax (Linum usitatissimum L.) and other species of the genus Linum L. Genet Resour Crop Evol 54:661–678
Diederichsen A, Hammer K (1995) Variation of cultivated flax (Linum usitatissimum L. subsp. usitatissimum) and its wild progenitor pale flax (subsp. angustifolium (Huds.) Thell.). Genet Resour Crop Evol 42:263–272
Everaert I, Rİek JD, Loose MD, Waes JV, Bockstaele EV (2001) Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Var Seeds 14:69–87
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.01: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fu YB (2005) Geographic patterns of RAPD variation in cultivated flax. Crop Sci 45:1084–1091
Fu YB, Allaby RG (2010) Phylogenetic network of Linum species as revealed by non-coding chloroplast DNA sequences. Genet Resour Crop Evol. doi:10.1007/s10722-009-9502-7
Fu YB, Peterson GW (2010) Characterization of expressed sequence tag derived simple sequence repeat markers for 17 Linum species. Botany (in press)
Fu YB, Peterson G, Diederichsen A, Richards KW (2002) RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet Resour Crop Evol 49:253–259
Galván MZ, Bornet B, Balatti PA, Branchard M (2003) Inter simple sequence repeat (ISSR) markers as a tool for the assessment of both genetic diversity and gene pool origin in common bean (Phaseolus vulgaris L.). Euphytica 132:297–301
Gill KS (1966) Evolutionary relationships among Linum species. Ph.D. Thesis, University of California, Riverside, USA
Gill KS (1987) Linseed. Indian Council of Agricultural Research, New Delhi
Godwin ID, Aitken EAB, Smith LW (1997) Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis 18:1524–1528
Hammer K (1986) Linaceae. In: Schultze-Motel J (ed) Rudolf Mansfelds Verzeichnis landwirtschaftlicher und gärtnerischer Kulturpflanzen. Akademie-Verlag, Berlin, pp 710–713 Bd. 2
Hamrick JL, Godt MJW (1997) Allozyme diversity in cultivated crops. Crop Sci 37:26–30
Heer O (1872) Über den Flachs und die Flachskultur im Altertum. Neujahrsblatt der Naturforschenden Gesellschaft Zürich 74:126
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Kulpa W, Danert S (1962) Zur Systematik von Linum usitatissimum L. [On the systematics of Linum usitatissimum L.]. Kulturpflanze. Beiheft 3:341–388
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Peakall R, Smouse PE (2005) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. The Australian National University, Canberra
Pharmawati M, Yan G, Finnegan PM (2005) Molecular variation and fingerprinting of Leucadendron cultivars (Proteaceae) by ISSR markers. Ann Bot 95:1163–1170
Rohlf FJ (1997) NTSYS-pc 2.1. Numerical taxonomy and multivariate analysis system. Exeter Software. Setauket, NY
SAS Institute Inc. (2004) The SAS system for windows V8.02. SAS Institute Incorporated. Cary, NC
Swofford DL (1998) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland
Tammes T (1928) The genetics of the genus Linum. Bibliogr Genet 4:1–36
Vromans J (2006) Molecular genetic studies in flax (Linum usitatissimum L.) Ph.D. Diss. Wageningen University, Wageningen, pp 41–59
Zietkiewicz E, Fafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
Authors would like to thank Mr. Dallas Kessler, Ms. Lorie Jones-Flory, and Ms. Kimberly MacKay for their technical assistance in sampling, planting, and screening the germplasm.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Uysal, H., Fu, YB., Kurt, O. et al. Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne Mill.) as revealed by ISSR markers. Genet Resour Crop Evol 57, 1109–1119 (2010). https://doi.org/10.1007/s10722-010-9551-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-010-9551-y