Abstract
Repositories of health records are collections of events with varying number and sparsity of occurrences within and among patients. Although a large number of predictive models have been proposed in the last decade, they are not yet able to simultaneously capture cross-attribute and temporal dependencies associated with these repositories. Two major streams of predictive models can be found. On one hand, deterministic models rely on compact subsets of discriminative events to anticipate medical conditions. On the other hand, generative models offer a more complete and noise-tolerant view based on the likelihood of the testing arrangements of events to discriminate a particular outcome. However, despite the relevance of generative predictive models, they are not easily extensible to deal with complex grids of events. In this work, we rely on the Markov assumption to propose new predictive models able to deal with cross-attribute and temporal dependencies. Experimental results hold evidence for the utility and superior accuracy of generative models to anticipate health conditions, such as the need for surgeries. Additionally, we show that the proposed generative models are able to decode temporal patterns of interest (from the learned lattices) with acceptable completeness and precision levels, and with superior efficiency for voluminous repositories.












Similar content being viewed by others
Notes
In this context, classification or predictive models should not be mingled with forecasting tasks, the unsupervised estimation of upcoming events for a time sequence, and regression tasks, the learning of parametric models to estimate a numeric attribute.
Given an alphabet \(\varSigma \), a generative string (sequence of contiguous events) is a probability distribution over \(\varSigma \) allowing substitutions with a certain noise probability \(\varepsilon \).
Assuming \(e_{j}(\sigma )\) to be the probability of emitting event’s value \(\sigma \) for a given \(x_j\) state, the entropy is given by: \(H(e_{j}) = -\varSigma _{\sigma } e_{j}(\sigma ) log(e_{j}(\sigma ))\).
Software available in http://web.ist.utl.pt/rmch/research/software.
http://www.heritagehealthprize.com/c/hhp/data (under a granted permission).
Software: http://www.philippe-fournier-viger.com/spmf/.
References
Abraham M, Ahlman J, Boudreau A, Connelly J, Evans D (2010) CPT 2011, Standard edn. American Medical Association Press, CPT / Current Procedural Terminology
Azuaje F (2011) Integrative data analysis for biomarker discovery. Omic Data Analysis for Personalized Medicine, Bioinformatics and Biomarker Discovery, pp 137–154
Baldi P, Brunak S (2001) Bioinformatics: The Machine Learning Approach. Adaptive Computation and Machine Learning. MIT Press, 2nd edition.
Batal I, Valizadegan Cooper, Hauskrecht M (2011). A pattern mining approach for classifying multivariate temporal data. In: IEEE BIBM, pages 358–365.
Baxter RA, Williams GJ, He H (2001) Feature selection for temporal health records. In PAKDD, pages 198–209, London, UK, UK. Springer-Verlag.
Bellazzi R, Ferrazzi F, Sacchi L (2011) Predictive data mining in clinical medicine: a focus on selected methods and applications. Wiley Interdisc. Rew. Data Mining and Knowledge Discovery 1(5):416–430
Bishop C (2006) Pattern Recognition and Machine Learning. Springer, Information Science and Statistics
Brand M (1999) Structure learning in conditional probability models via an entropic prior and parameter extinction. Neural Comput. 11(5):1155–1182
Brown M, Hughey R, Krogh A, Mian IS, Sjölander K, Haussler D (1993) Using dirichlet mixture priors to derive hidden markov models for protein families. In: 1st IC on Int. Sys. for Molecular Bio., pages 47–55. AAAI Press.
Bruno G, Garza P (2012) Temporal pattern mining for medical applications. In Data Min.: Found. and Int. Paradigms, volume 25 of ISRL, pages 9–18. Springer, Heidelberg.
Cao L, Ou Y, Yu PS, Wei G (2010) Detecting abnormal coupled sequences and sequence changes in group-based manipulative trading behaviors. In ACM SIGKDD, pages 85–94, New York, NY, USA. ACM.
Carreiro AV, Anunciação O, Carriço JA, Madeira SC (2011) Biclustering-based classification of clinical expression time series: A case study in patients with multiple sclerosis. In 5th International Conference on Practical Applications of Computational Biology & Bioinformatics (PACBB 2011), pages 229–239. Springer.
Choi K, Chung S, Rhee H, Suh Y (2010) Classification and sequential pattern analysis for improving managerial efficiency and providing better medical service in public healthcare centers. Healthc Inform Res. 16(2):67–76
Chudova D, Smyth P (2002) Pattern discovery in sequences under a markov assumption. In 8th ACM SIGKDD, KDD ’02, pages 153–162, New York, NY, USA. ACM.
Duan L, Street WN, Xu E (2011) Healthcare information systems: data mining methods in the creation of a clinical recommender system. Enterprise Information Systems 5(2):169–181
Eichler M (2012) Graphical modelling of multivariate time series. Probability Theory and Related Fields 153(1–2):233–268
Escobar G, Greene J, Scheirer P, Gardner M, Draper D, Kipnis P (2008) Risk-adjusting hospital inpatient mortality using automated inpatient, outpatient, and laboratory databases. Medical Care 46(3):232–239
Exarchos TP, Tsipouras MG, Papaloukas C, Fotiadis DI (2008) A two-stage methodology for sequence classification based on sequential pattern mining and optimization. Data Knowl. Eng. 66(3):467–487
Ge X, Smyth P (2000) Deformable markov model templates for time-series pattern matching. In ACM SIGKDD, pages 81–90, New York, NY, USA. ACM.
Guimarães G (2000) The induction of temporal grammatical rules from multivariate time series. In Proceedings of the 5th Int. Colloquium on Grammatical Inference: Algorithms and Applications, pages 127–140, London, UK. Springer-Verlag.
Guralnik V, Wijesekera D, Srivastava J (1998) Pattern directed mining of sequence data. In ACM SIGKDD, pages 51–57.
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The weka data mining software: an update. SIGKDD Explor. Newsl. 11(1):10–18
Henriques R, Antunes C (2014) Learning predictive models from integrated healthcare data: Extending pattern-based and generative models to capture temporal and cross-attribute dependencies. In System Sciences (HICSS), 2014 47th Hawaii International Conference on, pages 2562–2569.
Henriques R, Pina S, Antunes C (2013) Temporal mining of integrated healthcare data: Methods, revealings and implications. In SDM IW on Data Mining for Medicine and Healthcare, pages 52–60. SIAM.
Hu B, Chen Y, Keogh EJ (2013) Time series classification under more realistic assumptions. In: SDM, pages 578–586. SIAM.
Jacquemont S, Jacquenet F, Sebban M (2009) Mining probabilistic automata: a statistical view of sequential pattern mining. Mach. Learn. 75(1):91–127
Jain AK, Dubes RC (1988) Algorithms for clustering data. Prentice-Hall Inc, Upper Saddle River, NJ, USA
Laxman S, Sastry P, Unnikrishnan K (2005) Discovering frequent episodes and learning hidden markov models: A formal connection. IEEE TKDE 17:1505–1517
Letham B, Rudin C, Madigan D (2013) Sequential event prediction. Machine Learning 93(2–3):357–380
Li W, Han J, Pei J (2001) Cmar: Accurate and efficient classification based on multiple class-association rules. In ICDM, pages 369–376. IEEE CS.
Liu H, Motoda H (1998) Feature Selection for Knowledge Discovery and Data Mining. Kluwer Academic Publishers, Norwell, MA, USA
Mörchen F (2006) Time series knowledge mining. Wissenschaft in Dissertationen. Görich & Weiershäuser.
Murphy K (2002) Dynamic Bayesian Networks: Representation, Inference and Learning. PhD thesis, UC Berkeley, CS.
Nanopoulos A, Alcock R, Manolopoulos Y (2001) Information processing and technology. Feature-based classification of time-series data. Nova Science Publishers, Commack, NY, USA, pp 49–61
Norén G, Hopstadius J, Bate Star, Edwards I (2010) Temporal pattern discovery in longitudinal electronic patient records. Data Min. Knowl. Discov. 20(3):361–387
Pei J, Han J, Mortazavi-Asl B, Pinto H, Chen Q, Dayal U, Hsu M (2001) Prefixspan: Mining sequential patterns by prefix-projected growth. In ICDE, pages 215–224, Washington, DC, USA. IEEE CS.
Roverso D (2000) Multivariate temporal classification by windowed wavelet decomposition and recurrent neural networks. In ANS Int, Topical Meeting on NPICHMI
Sebastiani P, Ramoni M, Nolan V, Baldwin C, Steinberg M (2005) Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nature Genetics 37(4):435–440
Tseng V, Lee C-H (2009) Effective temporal data classification by integrating sequential pattern mining and probabilistic induction. Expert Sys. App. 36(5):9524–9532
Wan E (1990) Temporal backpropagation for fir neural networks. In IJC on Neural Networks, pages 575–580 vol. 1.
Yoo I, Alafaireet P, Marinov M, Pena-Hernandez K, Gopidi R, Chang J-F, Hua L (2012) Data mining in healthcare and biomedicine: A survey of the literature. Journal of Medical Systems 36(4):2431–2448
Acknowledgments
This work was supported by national funds through FCT - Fundaçã para a Ciência e a Tecnologia, under projects PEst-OE/EEI/LA0021/2013 (INESC-ID multiannual funding), PTDC/EIA-EIA/110074/2009 (D2PM), and the Ph.D. Grant SFRH/BD/75924/2011 to RH.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editors: Fei Wang, Gregor Stiglic, Ian Davidson and Zoran Obradovic.
Appendices
Appendix 1: Gathering and pre-processing collections of health records
1.1 Databases with integrated HRs
In the last decade, new patient-centric data sources emerged. Countries as United Kingdom and Netherlands, already track patients’ movements in the healthcare system across health providers, payors and suppliers . HR repositories are increasingly less fragmented, with appearing both cross-country and cross-players offerings, as provided by Cegedim and IMS. These repositories typically follow multi-dimensional and relational schema derived from electronic health records (McKesson, GE, PracticeFusion), imported health records (GoogleHealth, HealthVault) and claims (Ingenix, D2Hawkeye, CMS). Illustrating, GoogleHealth repositories provide an open interface to receive HRs from partners (often payors with interested in finding caregivers and discovering care tools) and to retrieve accessible personal HRs by users, lab and pharmacy systems. While some of these repositories capture more generalist healthcare views—e.g. HealthVault goal is to facilitate the exchange of health information among patients, care givers and service providers to support health decisions—, others repositories focus on more specific health views—e.g. Continua monitors the health and care needs of chronic diseased patients. Enablers of more complete HR repositories include the privileged access to device-related patient data and in-home transmitters, and emerging integrated connections across a wide-variety of systems, such as with patient communities (Alere, Pharos, SilverLink, WebMD, HealthBoards), consumer reports (Anthem, vimo, hospitalcompare), content aggregators (Walters Kluer, Reed Elsevier, Thomson) and online worksite healthcare (iTrax, webConsult).
1.2 Pre-processing HR databases
The health records from the surveyed repositories are typically integrated through a multi-dimensional or relational schema. A multi-dimensional data structure is defined by a set of dimensions and a central fact. The central fact contains one foreign key for each dimension and a set of measures. Relational schema can be easily mapped as a multi-dimensional schema where the entity-relationship table capturing the HRs (either electronic, imported or derived from claims) is seen as the central fact table, and its linked tables as the dimensions. However, due to the high number of monitored healthcare attributes related with lab results, prescriptions, treatments and diagnostics, each HR entry would define a highly sparse set of measures. To tackle this problem, all the possible HR measures are replaced by a tuple that defines the name and domain of the attribute (as a foreign key to the attribute dimension) and the monitored values. Figure 13 illustrates an HR-centered multi-dimensional database. For simplicity sake, in this work we map numeric domains as ordinal domains, complex data as a complex set of categorical values and we ignore free-text. From these pre-processed entries, obtaining a collection of events follows a two-stage process. First, the type of attributes is recorded and each pair (attribute, value) is fixed. Second, two special dimensions, the date and split dimensions, are used to retrieved the remaining fields associated with an event. The time dimension is used to obtain the timestamp based on the time of occurrence of each monitored entry. The split dimension (commonly the patient) is used to obtain the instance identifier (patientID) using the primary keys of this dimension. The number of instances in the training dataset is given by the number of these primary keys.
Appendix 2: Architectural components
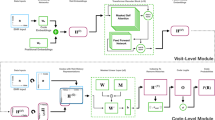
Consider a variant of a left-to-right architecture, where main and insert states only emit events delimited by two states that can only emit the delimiter symbol with transitions to self-looping states. This architecture, referred as CoIA (Co-occuring Items Architecture) and illustrated in Fig. 14, captures co-occurring events that are frequently observed for a specific medical condition. In this way, CoIA is able to model cross-attribute dependencies as co-occurring events that may be associated with different attributes of interest. A transition to the initial state can be placed to consider recurrence within a sequence of event-sets. Optionally, an end state can be linked to the architecture. This guarantees that, at least, one set of co-occuring events is used to learn the left-to-right emissions. The end state can be implemented by adding a dedicated ending event at the end of each sequence of event-sets. An alternative to CoIA is to consider a fully-interconnected architecture.
Definition 6
CoIA(i) is an architectural component \((T,E)\) over \(\{x_{Ai}, pBlock_i(1),\) \(\ldots ,pBlock_i(L),\) \(x_{2L+1} \}\) states where \(L > \overline{\hat{p}}.l\). The transition probabilities are \(t_{(Ai)(Ai)}\) = 1-\(\eta \), \(t_{(Ai)(11)}=\eta \alpha \), \(t_{(Ai)(21)}=\eta \beta \), \(t_{(Ai)(31)}=\eta \gamma \), \(t_{(1L)(Ai+1)} \in \{\eta \alpha ,0\} \ne t_{(1L)(Ai)} \in \{\eta \alpha ,0\}\) , \(t_{(2L+1)(Ai+1)} \in \{\eta \beta ,0\}\) \(\ne t_{(2L+1)(Ai)} \in \{\eta \beta ,0\}\), \(t_{(3L)(Ai+1)} \in \{\eta \gamma ,0\} \ne t_{(3L)(Ai)} \in \{\eta \gamma ,0\}\) , where \(\beta =1\)-\(\alpha \)-\(\gamma \) and \(\eta =\frac{l}{n\times l}\) for the no recurrence option (when \(t_{(pBlock_i(L))(Ai)}\) = 0) and \(\eta =\frac{1}{l}\) when allowing recurrence. Emissions are defined as \(\forall _{\sigma \in \varSigma }\) \(e_{Ai} (\sigma )=\frac{1-\kappa }{\mid \varSigma \mid }\) and \(e_{Ai} (\$)=\kappa \), where \(\kappa \) is either the fraction number of delimeters from all symbols (no recurrence) or 1 (recurrence).
Consider now the proposed Itemset-Precedences Architecture (IPA) illustrated in Fig. 15. With this architecture we can model precedences between events. Two aspects of IPA should be noticed. First, insert states are used to remove events that do not occur frequently for a particular medical condition. Second, each state dedicated to emit delimiters has a transition to a self-looping state in order to allow for gaps between events. In this way, we transit from co-occuring events to precedences. However, besides not capturing co-occuring events, IPA suffers from another drawback. Since there is no guarantee that most of the input sequences will reach state \(x_N\), the significance of precedences decoded from the first portions of the path is greater than from those decoded from the last portions. Similarly to CoIA, we can consider an IPA variant that includes deletions for minimizing this problem and allowing for a well-distributed significance of emissions across the main path.
Appendix 3: Pattern-based predictive models
Under the mapping proposed in Sect. 3.1, the existing methods for the analysis of itemset sequences can be extended to deal with sequences of event-sets, and their output used to guide and shape the target predictive models. The most common task in this context is the discovery of sequential patterns to mine frequent precedences and co-occurrences. Sequential patterns discovered over the target temporal structure are able to include events associated with multiple healthcare attributes of interest.
Different strategies have been proposed for the use of temporal patterns for classification (Exarchos et al. 2008; Nanopoulos et al. 2001). However, they are only prepared to capture frequent precedences and co-occurrences and, therefore, are not able to consider temporal distances between events, which is a critical requirement for the definition of predictive models. Additionally, they have been developed in the scope of genomic studies and multivariate time series analysis, and, consequently, the argued levels of performance no longer remain valid for repositories of health-records. For these reasons the authors Henriques and Antunes (2014) proposed a new pattern-based classifier, referred as P2MID (Pattern-based Predictive Models from Integrated Data).
The behavior of the P2MID classifier can be described according to its training and testing stages. In the training stage a discriminative model is defined in three steps. First, a set of time-enriched sequential patterns is generated for each medical condition.
Definition 7
A time-enriched sequential pattern is a sequential pattern observed for specific time interval \([\varphi _i,\varphi _f]\). A time-enriched sequential pattern is subset of another, \(a\subseteq b\), if it is both a subsequence and its time interval is contained in \(a\) time range (\(\varphi _{a_i} \le \varphi _{b_i}\wedge \varphi _{a_f} \ge \varphi _{b_f}\)).
Considering the illustrative set, \(\{(ac)\emptyset db, (ac)d\emptyset \emptyset ,\) \((ac)\emptyset \emptyset (bd)\}\), and a minimum support \(\theta =2\), \((ac)d\) is a simple sequential pattern, while \(\{(ac)d\}[\varphi _i=0,\) \(\varphi _f=2]\) or \(\{db\}[\varphi _i=3,\varphi _f=4]\) are illustrative time-enriched sequential patterns for a granularity \(\delta =1\). P2MID computes these temporal patterns by fixing multiple temporal aggregations (\(\delta \in \{1,2,\ldots \}\)) followed by the discovery of co-occurrences for coarser-grained aggregations under a penalization factor to benefit the discovery patterns that occur for small time intervals.
Second, the confidence of each pattern in relation to a particular medical condition is evaluated to compose rules of the form \(s\Rightarrow y\), where \(s\) is the temporal pattern and \(y\in Y\) is the condition (class).
Third, and similarly to CMAR (Li et al. 2001), these rules are inserted in a tree structure if (1) the \(\chi ^2\) test over the rule is above a specified \(\alpha \)-significance level, and if (2) the tree does not contain a rule with higher priority. Since CMAR is not able to deal with temporal patterns (only with frequent itemsets), we propose a new priority criterion. A rule \(R_1:s_1\Rightarrow y\) has priority over \(R_2:s_2\Rightarrow y\) if \(s_1\subseteq s_2\) or if:
Finally, the tree is pruned based on the computed priorities.
This tree defines the discriminative pattern-based model, which is a simple ordered set of tuples (pattern \(s\), class \(y\), weight \(\beta \)).
In the testing stage of P2MID, this discriminative model is used to classify a specific patient by identifying the closest temporal patterns and relying on their matching score for the target conditions. The strength of each condition is calculated by computing the weighted-\(\chi ^2\) across all the rules \(s\Rightarrow y\) that satisfy a matching criterion between the pattern \(s\) and the testing instance.
with \(MCS=(min(sup(s),sup(y))-sup(s)sup(y)/N)^2 \times N \times e\), where:
Matching occurs if the pattern is observed for the testing instance within the specified time frame or within the specified duration but for different time partitions (time shift condition). For this case, the number of shifted partitions is used to penalize the rule score.
Finally, the strongest condition, \(y\in Y\), is delivered (deterministic output) or, alternatively, the computed strength for each class (probabilistic output).
1.1 Results
In Fig. 16, the impact of adopting time-enriched sequential patterns versus simple sequential patterns is evaluated. The difference in performance is statistically significant (at \(\alpha \) = 2 %). First, the proposed discriminative models tend to score preferentially patterns occurring near the time period under prediction. Also, the allowance of temporal shifts under a penalization factor during the testing stage offers a time-dependent informative context for classification. Contrasting, simple sequential patterns cannot offer temporal guarantees, and, therefore, the influence of both recent and old events to discriminate the class under prediction is not clearly differentiated. Second, the time partitioning strategy allows to deal with arbitrary high levels sparsity by choosing adequate granularity levels with impact on the degree of precedences vs. co-occurrences. Finally, the integration of both simple and time-enriched sequential patterns was observed to slightly increase the overall levels of accuracy. This can be explained by the inclusion of precedences (modeled by simple sequential patterns) with larger time frames.
1.2 Illustrative case
Consider the illustrative task of predicting the need of a specific treatment \(T1\) for the upcoming quarter from healthcare data monitored along 2 years. Consider the presence of 15 clinical procedures (\(T\)1-15), 10 major health conditions (\(D\)1-10), 20 lab-test assessments (\(L\)1-20) and 15 categories of prescriptions (\(P\)1-15). Let us assume that, under a selected month granularity, the learned P2MID predictor has the following top 5 rules: \(\{(D5L8P3)P3\}[20,24] \Rightarrow T1\) (confidence \(c\) = 97 % and priority score \(\beta \) = 37), \(\{T1\}[12,24]\) \(\Rightarrow \lnot T1\) (\(c\) = 91 %,\(\beta \) = 35), \(\{(D4L8P2)P2\}[16,22] \Rightarrow T1\) (\(c\) = 89 %,\(\beta \) = 34), \(\{L6P7\}[18,24] \Rightarrow \lnot T1\) (\(c\) = 78 %,\(\beta \) = 31) and \(\{P4(D4D5)\}[14,20] \Rightarrow T1\) (\(c\) = 8 %,\(\beta \) = 28). Understandably, a patient under prediction with the following sequence of event-sets, \(\{(T2P8)(L2P8)(D5P3)P3\emptyset (L8D4P2\) \(P3)(P2P3)\}\)[18,24], is likely to be classified as a candidate for \(T1\) treatment. Note that matching criteria is enough expressively to consider temporal misalignments and event gaps.
Appendix 4: Decoding arrangements of events from HMM architectures
Robust and efficient methods can be defined to decode the most probable arrangements of events from the learned HMM lattices. For this purpose, we propose the graph mining method described in Algorithm 1.

Algorithm 1 relies on key properties. First, the use of the anti-monotonic property to prune paths. Second, the use of probability thresholds that traduces the criterion that defines whether a pattern is or not frequent. This threshold can be weighted by the length of the pattern. This avoids a biased orientation towards small sequential patterns. Finally, the use of a minimum probability threshold for emissions and transitions. This property is critical to guarantee an heightened efficiency of the graph mining method and to avoid the output of non-frequent arrangements by selecting only the most probable event emissions by state (convergence criterion).
Appendix 5: Detailed analysis of the behavior of the proposed HMMs
An initial study of the behavioral specificities of the proposed HMMs was provided in Sect. 4.2. Below, we complement this analysis by providing the experimental details required for the reproducibility of the generated datasets and additional results for an in-depth study of the target generative models. Based on the observed arrangement of HRs for the heritage database, we fixed the input average length of maximal patterns of the IBM tool generator as 4 and the average maximal frequent transactions as 2. The values for different sequential patterns and transactional patterns were set to 1.000 and 2.000, respectively. The remaining parameters were provided in Sect. 4.2. The default setting generates near 10.000 sequential patterns (sets of events satisfying ordering constraints) for a support of 1 % (with the majority of them having more than 5 events/items), and more than 400 sequential pattens for a support of 4 %.
Below we study the ability of the proposed HMMs in modeling the planted arrangement of events (sequential patterns). For this goal, we use completeness and precision metrics in order to study how the arrangements of events decoded from these generative models (according to Appendix ) match the planted arrangements (corresponding to the output of deterministic approaches).
1.1 Completeness
In this analysis, we want to guarantee that, at least, all the arrangements of events with support above 4 % are captured. Figure 17 illustrates the completeness of the proposed architectures. Note that an increase of support to 6 % results in approximated levels of 100 % for all the architectures across datasets. Density is given by the average number of events divided by the cardinality of domain values, \(\frac{n\times l}{\mid \varSigma \mid }\). Two major observations result from the analysis of completeness levels. First, the adoption of multi-path, integrative and fully-interconnected architectures achieve a good completeness since they can focus on different subsets of probable emissions along the alternative architectural paths. Second, the levels of completeness degrade for higher densities and lengthy number of events. This is a natural result of the explosion of sequential patterns discovered by deterministic approaches under such hard settings. These large outputs become hardly coverable by the proposed compact architectures. A viable solution is to rely on integrated architectures with a higher number of paths. An experiment with an architecture composed of six SPA paths was able to hold completeness levels above 96 % for all the adopted settings.
1.2 Precision
We consider a decoded arrangement of events to be deterministically frequent if it has a support higher than 1.5 %. Figure 18 illustrates the precision of the proposed architectures, that is, the fraction of decoded patterns deterministically frequent. Note that a decrease of support to 0.8 % results in an approximated levels of 100 % for all the architectures across datasets. A key observation can be derived from this analysis: generative approaches hold high levels of precision (\(>\)90 %) across the majority of data settings. There is a slight decrease of precision for very sparse sequences of event-sets (average of 2 events per time partition) or for small sequences (average of 4 time partitions) since the number of deterministic patterns is smaller than the average number of patterns able to decode from the adopted architectures. Also, there is a slight decrease of precision for large average number of events per partition that mainly results from an accumulative error associated with the decoding of larger patterns, that is also explained by the size constraints for co-occurring events adopted for SPA architectures.
The greatest challenge for achieving a high precision when adopting generative models seems to be related with a high cardinality of domain values, \(\mid \varSigma \mid \). The observed decrease in precision with increased cardinality is not only explained by a reduced set of deterministic patterns (potentially smaller than the decoded set) but also by an intrinsic difficulty to guarantee the convergence towards a reduced set of emissions.
1.3 Efficiency
The comparison of efficiency for the alternative architectures against PrefixSPAN (Pei et al. 2001), one of the most efficient deterministic SPM algorithms, is illustrated in Fig. 19. Under 1 % support threshold, the number of deterministic frequent patterns vary between 1.000 patterns (smaller datasets) to near 100.000 patterns (denser and larger datasets) across settings. Three major observations can be retrieved from the efficiency analysis. First, generative approaches are particularly suitable over datasets with lower cardinality of values against deterministic approaches, whose performance rapidly deteriorates for densities above 10 %.
Contrasting, the performance of the generative approaches does not significantly change with varying densities. This is explained by a double-effect: learning convergence deteriorates with an increased density, but this additional complexity is compensated by a higher efficiency per iteration since there is a significantly lower number of emission probabilities to learn per state. Second, generative models scale better with either an increase number of time partitions or events per time partition than PrefixSPAN. Under the default settings, PrefixSPAN is efficient for sequence lengths with less than 15 partitions. Finally, fully-interconnected and LRA are the most efficient architectures due to their structural simplicity. However, this efficiency gain has a precision cost as the high number of self-loops turn the decoded arrangements of events more prone to local errors. The remaining architectures have higher complexity, although are still considerably preferable over deterministic alternatives for hard settings.
Rights and permissions
About this article
Cite this article
Henriques, R., Antunes, C. & Madeira, S.C. Generative modeling of repositories of health records for predictive tasks. Data Min Knowl Disc 29, 999–1032 (2015). https://doi.org/10.1007/s10618-014-0385-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-014-0385-7