Abstract
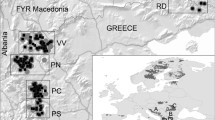
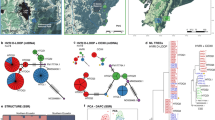
The formerly large, continuous brown bear population of the Carpathians has experienced a radical decrease in population size due to human activities which have resulted in splitting the population into the larger Eastern Carpathian and the smaller Western Carpathian subpopulations. In the Western Carpathians, brown bears came close to extinction at the beginning of 1930s, but thanks to both conservation and management efforts the bear population has begun to recover. In contrast, the Eastern Carpathian subpopulation in Romania has never dropped below 800 individuals, potentially preserving the original amount of genetic variation. In this paper we present results of a genetic study of brown bear subpopulations distributed in the Slovak and Romanian sections of the Carpathians using 13 nuclear microsatellites. The documented level of genetic differentiation between the Western and Eastern Carpathian subpopulations reflects the isolation which lasted almost 100 years. Furthermore, the existence of two, different, genetic clusters within the Western Carpathians despite close geographic proximity indicates that human-caused fragmentation and isolation have resulted in significant genetic divergence. Although the subpopulations display an indication of genetic bottleneck, the level of genetic diversity is within the range commonly observed in different brown bear populations. The results presented here point out the significance of human exploitation to the population structure of this large carnivore species. Future management efforts should be aimed at securing and restoring the connectivity of forested habitats, in order to preserve the genetic variation of the Carpathian brown bear subpopulations and to support the gene flow between them.




Similar content being viewed by others
References
Adams JR, Waits LP (2007) An efficient method for screening faecal DNA genotypes and detecting new individuals and hybrids in the red wolf (Canis rufus) experimental population area. Conserv Genetics 8:123–131
Anonymous (2007) Management and action plan for the bear population in Romania. Ministry of Agriculture, Forestry and Rural Development, Ministry of Environment and Water Management. Bucureşti, pp 1–79. Available at http://www.icas.ro/DOCS/Bear%20Management%20Plan.pdf
Aurelle D, Cattaneo-Berrebi G, Berrebi P (2002) Natural and artificial secondary contact in brown trout (Salmo trutta L.) in the French western Pyrenees assessed by allozymes and microsatellites. Heredity 89:171–183
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996–2004) genetix 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier
Bellemain E, Taberlet P (2004) Improved noninvasive genotyping method: application to brown bear (Ursus arctos) faeces. Mol Ecol Notes 4:519–522
Bierne N, Borsa P, Daguin C et al (2003) Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol Ecol 12:447–461
Caro TM, Laurenson MK (1994) Ecological and genetic factors in conservation: a cautionary tale. Science 263:485–486
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crnokrak P, Roff DA (1999) Inbreeding depression in the wild. Heredity 83:260–270
Dahle B, Swenson JE (2003) Home ranges in adult Scandinavian brown bears (Ursus arctos): effect of mass, sex, reproductive category, population density and habitat type. J Zool 260:329–335
Di Rienzo A, Peterson AC, Garza JC et al (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Ehrich D, Gaudeul M, Assefa A et al (2007) Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Mol Ecol 16:2542–2559
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Evett I, Weir B (1998) Interpreting DNA evidence: statistical genetics for forensic scientists. Sinauer Associates, Sunderland
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online 1:47–50
Finďo S, Skuban M, Koreň M (2007) Brown bear corridors in Slovakia. Identification of critical segments of the main road transportation corridors with wildlife habitats. Carpathian Wildlife Society, Zvolen, pp 1–68
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Glaubitz JC (2004) Convert: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310
Gouin N, Grandjean F, Souty-Grosset C (2006) Population genetic structure of the endangered crayfish Austropotamobius pallipes in France based on microsatellite variation: biogeographical inferences and conservation implications. Freshw Biol 51:1369–1387
Hartl GB, Hell P (1994) Maintanence of high-levels of allelic variation in spite of a severe bottleneck in population size–the brown bear (Ursus arctos) in the Western Carpathians. Biodivers Conserv 3:546–554
Hell P, Slamečka J (1999) Bear in Slovak Carpathians and in the world [in Slovak]. PaRPRESS, Bratislava
Ionescu O (1999) Status and management of the brown bear in Romania. In: Servheen C, Herrero S, Peyton B (eds) Bears: status survey and conservation action plan. International Union for the Conservation of Nature and Natural Resources, Switzerland, pp 93–96
Jakubiec Z (2001) The Brown bear Ursus arctos L. in the Polish part of the Carpathians (in Polish). Polska Akademia Nauk, Instytut Ochrony Przyrody, Studia Naturae 47
Jiménez JA, Hughes KA, Alaks G, Graham L, Lacy RC (1994) An experimental study of inbreeding depression in a natural habitat. Science 266:271–273
Jombart T, Pontier D, Dufour AB (2009) Genetic markers in the playground of multivariate analysis. Heredity 102:330–341
Karamanlidis AA, Drosopoulou E, Hernando MG, Georgiadis L, Krambokoukis L, Pllaha S, Zedrosser A, Scouras Z (2010) Noninvasive genetic studies of brown bears using power poles. Eur J Wildl Res 56:693–702
Kocijan I, Galov A, Ćetković H, Kusak J, Gomerčić T, Huber D (2011) Genetic diversity of Dinaric brown bears (Ursus arctos) in Croatia with implications for bear conservation in Europe. Mammalian Biology, doi:10.1016/j.mambio.2010.12.003
Kohn M, Knauer F, Stoffella A, Schröder W, Pääbo S (1995) Conservation genetics of the European brown bear–a study using excremental PCR of nuclear and mitochondrial sequences. Mol Ecol 4:95–103
Lacy RC (1993) Impacts of inbreeding in natural and captive populations of vertebrates: implications for conservation. Perspect Biol Med 36:480–496
Lande R (1988) Genetics and demography in biological conservation. Science 241:1455–1460
Manoukis NC (2007) Formatomatic: a program for converting diploid allelic data between common formats for population genetic analysis. Mol Ecol Notes 7:592–593
Miller CR, Joyce P, Waits LP (2002) Assessing allelic dropout and genotype reliability using maximum likelihood. Genetics 160:357–366
Paetkau D, Strobeck C (1994) Microsatellite analysis of genetic variation in black bear populations. Mol Ecol 3:489–495
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354
Paetkau D, Waits LP, Craighead L, Clarkson P, Strobeck C (1998) Variation in genetic diversity across the range of North American brown bears. Conserv Biol 12:418–429
Paetkau D, Slade R, Burden M, Estoup A (2004) Genetic assignment methods for direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mole Ecol 13:55–65
Paszlavsky J (1918) Classis Mammalia. Fauna regni Hungariae, Mammalia. Regia Societas Scientia Naturalium Hungarica, Budapest, 43 pp
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Piry S, Alapetite A, Cornuet JM et al (2004) Geneclass2: a software for genetic assignment and first-generation migrant detection. J Heredity 95:536–539
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005) Genotyping errors: causes, consequences and solutions. Nat Rev Genetics 6:847–859
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for structure software: Version 2.2. Avaiable at http://pritch.bsd.uchicago.edu/software/structure2_2.html
Ralls K, Ballou JD, Templeton A (1988) Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv Biol 2:185–193
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201
Rosenberg NA, Pritchard JK, Weber JL et al (2002) Genetic structure of human populations. Science 298:2381–2385
Saccheri I, Kuussaari M, Kankare M et al (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392:491–494
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Laboratory Press, New York
Servheen C (1990) Status and conservation of the bears of the world. International conference on bear research and management No. 2, 34 pp
Shaw PW, Pierce GJ, Boyle PR (1999) Subtle population structuring within a highly vagile marine invertebrate, the veined squid Loligo forbesi, demonstrated with microsatellite DNA markers. Mol Ecol 8:407–417
Swenson JE, Wabakken P, Sandegren F, Bjärvall A, Franzén R, Söderberg A (1995) The near extinction and recovery of brown bears in Scandinavia in relation to the bear management policies of Norway and Sweden. Wildl Biol 1:11–25
Swenson JE, Dahle B, Gerstl N, Zedroser A (2000) Action plan for conservation of the brown bear in Europe. Convention on the Conservation of European wildlife and natural habitats (Bern Convention), nature and environment, Nr. 114. Council of Europe Publishing, Strasbourg
Taberlet P, Bouvet J (1994) Mitochondrial DNA polymorphism, phylogeography, and conservation genetics of brown bear (Ursus arctos) in Europe. Proc R Soc Lond Ser B 255:195–200
Taberlet P, Luikart G (1999) Non-invasive genetic sampling and individual identification. Biol J Linn Soc 68:41–55
Taberlet P, Swenson JE, Sandegren F, Bjärvall A (1995) Localization of a contact zone between two highly divergent mitochondrial DNA lineages of brown bear Ursus arctos in Scandinavia. Conserv Biol 9:1255–1261
Taberlet P, Camarra JJ, Griffin S et al (1997) Noninvasive genetic tracking of the endangered Pyrenean brown bear population. Mol Ecol 6:869–876
Tobiáš J (1933) Bears in Turiec region [in Slovak]. Lovec 6:4–8
Valière N (2002) Gimlet: a computer program for analyzing genetic individual identification data. Mol Ecol Notes 2:377–379
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol 4:535–538
Waits L, Taberlet P, Swenson JE, Sandergren F, Franzén R (2000) Nuclear analysis of genetic diversity and gene flow in the Scandinavian brown bear (Ursus arctos). Mol Ecol 9:421–431
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Zachos FE, Otto M, Unici R, Lorenzini R, Hartl GB (2008) Evidence of a phylogeographic break in the Romanian brown bear (Ursus arctos) population from the Carpathians. Mammalian Biol 73:93–101
Žuffa A (1932) Game in Tatras (in Slovak). Lovec 21:1–2
Acknowledgements
This study was supported by Slovak Research and Development Agency through financial support APVV-18-032105. Authors would like to thank to State Nature Conservancy of the Slovak Republic for providing the tissue samples of legally and accidentally killed animals and sample collectors from Slovakia and Romania for assistance with sampling. Also we would like to thank to D. Krajmerová, G. Baloghová for help with laboratory work; to D. Gömöry for advice with statistical analyses, and to L. Waits, T. Skrbinšek and P. Kaňuch for their valuable comments on the early version of this manusript. Thanks are due to Dr. E. Ritch-Krč for improving English.
Author information
Authors and Affiliations
Corresponding author
Appendix 1
Rights and permissions
About this article
Cite this article
Straka, M., Paule, L., Ionescu, O. et al. Microsatellite diversity and structure of Carpathian brown bears (Ursus arctos): consequences of human caused fragmentation. Conserv Genet 13, 153–164 (2012). https://doi.org/10.1007/s10592-011-0271-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-011-0271-4