Abstract
Several recent studies have produced comparative maps of genes on amniote sex chromosomes, revealing homology of gene content and arrangement across lineages as divergent as mammals and lizards. For example, the chicken Z chromosome, which shares homology with the sex chromosomes of all birds, monotremes, and a gecko, is a striking example of stability of genome organization and retention, or independent acquisition, of function in sex determination. In other lineages, such as snakes and therian mammals, well conserved but independently evolved sex chromosome systems have arisen. Among lizards, novel sex chromosomes appear frequently, even in congeneric species. Here, we review recent gene mapping data, examine the evolutionary relationships of amniote sex chromosomes and argue that gene content can predispose some chromosomes to a specialized role in sex determination.
Similar content being viewed by others
Sex chromosomes in amniotes
Sex determination, the process by which development is directed down the male or female pathway, has been the subject of philosophical interest for millennia (Mittwoch 2000) and rigorous enquiry in more recent times (Bellott and Page 2010; Mank 2011; Ming et al. 2011; Ito et al. 2011). Even when the focus is narrowed to amniotes (comprising reptiles, birds, and mammals), an astonishing variety of mechanisms determine sex, suggesting frequent evolution and turnover of sex chromosomes as well as sex-determining modes.
In mammals (and some turtles and lizards) with genotypic sex determination (GSD), heterogametic males segregate heteromorphic sex chromosomes called X and Y, the latter of which is often heterochromatic and gene poor. Females have an XX constitution. In snakes and birds (as well as some turtles and lizards), males are homogametic, and females produce gametes with different sex chromosomes, called Z and W. In some GSD amniotes (e.g., pythons, ratite birds, and many lizards), sex chromosomes are so similar in morphology that the heterogametic sex cannot be determined using standard cytogenetic techniques (Ezaz et al. 2005, 2009b; Gamble 2010). In several reptiles, sex is determined not by chromosomal contributions of the parents, but by the influence of ambient temperature on the egg at a critical period in embryonic development, known as temperature-dependent sex determination (TSD) (Bull 1983; Charnier 1966). Orthologs of genes on the sex chromosomes of GSD species are borne by autosomes in TSD species so do not segregate with sex. Although the trigger for determining sex is so variable, these genetic or environmental factors initiate a complex regulatory signal cascade that is remarkably similar in all taxa, involving a network of genes and hormones that ultimately—and reliably—produces either male or female offspring (Georges et al. 2010).
In all but a few exceptional mammals, a single dominant gene on the Y chromosome (SRY) is responsible for the primary signal that directs sexual differentiation down the male pathway (Koopman et al. 1991; Sinclair et al. 1990). In birds, male development is effected, not by presence or absence of a dominant gene, but by copy number (dosage) of the Z-borne DMRT1 (Smith et al. 2009), as well as an unknown cell autonomous factor (Clinton et al. 2011). A thermosensitive gene may have captured initiation in reptiles with TSD (Rhen et al. 2011), or sex may be determined more by overall displacement of the sex differentiation pathway by temperature early in development (typically the middle third) while the differentiating gonad is still bipotential (Georges et al. 2010). Sex determination by genotype and temperature are not mutually exclusive modes, as genotype and environment can interact to determine sex in some species (Ospina-Álvarez and Piferrer 2008; Quinn et al. 2007; Radder et al. 2008).
The determination of sex by a constitutional difference in the genotype of males and females often, but not always, leads to marked differences in morphology and gene content of the chromosome pair responsible for sex determination (Bergero and Charlesworth 2009; Graves 2008; Perrin 2009). These differences derive partly from inefficient selection on the non-recombining sex-specific Y or W chromosome, and partly from the different proportion of evolutionary time that the X and Y, or the Z and W, spend in males and females (reviewed by Mank et al. (2010), Bachtrog (2006), Bellott and Page (2010), and Bergero and Charlesworth (2009)).
The diverse mechanisms of sex determination in amniotes have raised longstanding questions about sex chromosome evolution: Why are some sex chromosomes so well conserved and others so changeable (Ezaz et al. 2009b; Nanda et al. 2008; Ohno 1967)? Are amniote sex chromosomes all derived from a common ancestral autosome (Bellott et al. 2010; Smith and Voss 2007)? Are some chromosomes better suited to a role in sex determination than others? Graves and Peichel (2010) asked whether similarity of sex chromosomes in divergent taxa reflect “shared ancestry or limited options,” and concluded that, at least in some cases, the same genomic region had independently acquired a role in sex determination. Much recent work has focused on the remodeling of sex chromosomes after a pair of autosomes takes on that role (e.g., Bellott et al. 2010; Bergero and Charlesworth 2009; Potrzebowski et al. 2008). Instead, we examine the idea that some sex chromosomes were derived from autosomes with preexisting features that lend them to such a role.
Sex chromosomes of birds, snakes, and therian mammals are nonhomologous
Sex chromosomes are ubiquitous in mammals, birds, and snakes and are known in some turtles and many lizards (Fig. 1). More than 40 years ago, Susumu Ohno speculated that despite their opposite systems of heterogamety, mammal, bird, and snake sex chromosomes had arisen from the same ancestral autosomal pair because of their common involvement in sex determination (Beçak et al. 1964; Ohno 1967).
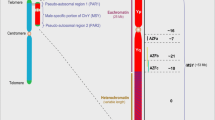
Phylogeny of tetrapods and amniote sex chromosome homologies. a A consensus tree of tetrapod relationships, indicating the distribution of sex determination modes and sex chromosome systems. Branch lengths are proportional, with divergence dates from Bininda-Emonds et al. (2007) and Hedges and Kumar (2009). Only representative taxa are shown to give some indication of the depth of divergence in the major groups. The uncertain placement of turtles (Testudines) is indicated by a dotted line (Hugall et al. 2007; Werneburg and Sánchez-Villagra 2009). MYA millions of years ago, XY male heterogamety, ZW female heterogamety, TSD temperature-dependent sex determination. b Generalized representation of chromosome and linkage group homologies of the chicken Z chromosome (green); the turtle Pelodiscus sinensis Z chromosome (magenta); the snake Elaphe quadrivirgata Z chromosome (yellow and cyan); and the human X chromosome (blue & red). There is little evidence of an ancestral syntenic association of snake, bird, and mammal sex chromosomes, but the Pelodiscus Z almost certainly derives from the same ancestral chromosome that gave rise to the Anolis X, the gecko and bird Z, and platypus X5. Finer scale gene mapping in reptiles and amphibians may reveal as yet undetected ancestral chromosomal associations. Mapping and painting data are from http://ensembl.org and Alfoldi et al. (2011), Bellott et al. (2010), Ezaz et al. (2009a), Hellsten et al. (2010), Kawagoshi et al. (2009), Kawai et al. (2009, 2007), Matsubara et al. (2006), Matsuda et al. (2005), Mikkelsen et al. (2007), O’Meally et al. (2009), Shetty et al. (1999), Srikulnath et al. (2009a, b), and Uno et al. (2008). LG linkage group, μ unidentified microchromosome
However, comparative gene mapping and, more recently, complete sequence analysis of several species of birds and mammal sex chromosomes have dispelled any notion of homology between the therian X and avian Z (Bellott et al. 2010; Mikkelsen et al. 2007; Nanda et al. 2002). Like the mammalian X (Ohno 1967; Rodríguez Delgado et al. 2009; Ross et al. 2005), synteny of avian Z-borne genes is highly conserved across species (Nanda et al. 2008; Shetty et al. 1999). In therian mammals, the chicken Z corresponds to autosomal regions: parts of chromosomes 5, 9, and 18 in humans and to parts of opossum chromosomes 3 and 6 (Figs. 1b and 2). The human X corresponds to parts of chicken chromosomes 1 and 4. None of the 1,000 or so genes on the chicken Z lies on the X in therian mammals and vice versa for the approximately 1,100 genes on the human X (Bellott et al. 2010).
Comparative map of chicken, ratsnake, and human sex chromosomes. Sex chromosomes are ubiquitous in the amniote groups represented here, though they are not homologous by gene content. The snake Z is composed of genes found on chicken chromosomes 2 (yellow) and 27 (cyan). The chicken Z (green) is equivalent to parts of chromosome 2 in the ratsnake and parts of human chromosomes 5, 9, and 18. The human X is orthologous to parts of chicken chromosomes 1 (red) and 4 (blue). Mapping data are from http://ensembl.org and Bellott et al. (2010), Matsubara et al. (2006), and Matsuda et al. (2005)
Comparative mapping of 11 Z-borne genes across three divergent snake families (a python, colubrid, and viper) shows that they share a conserved Z chromosome that is clearly not homologous with the bird Z (Fig. 2) (Matsubara et al. 2006). The chicken Z is orthologous to parts of chromosome 2 in the four-lined ratsnake, Elaphe quadrivirgata. The snake Z bears orthologs of genes on chicken chromosomes 2p and 27. To date, 29 genes mapped to the snake Z have orthologs on these two chicken chromosomes (K. Matsubara, personal communication), indicating that this chromosome arose by fusion of ancestral segments represented by chicken chromosomes 2p and 27 after a translocation between Z and 2 in sauropsid ancestor (O’Meally et al. 2010). Synteny of chicken 2 and Z genes is conserved in monotremes (Rens et al. 2007), suggesting that the translocation occurred in an amniote ancestor.
Independent and multiple origins
Genome sequencing and gene mapping have confirmed that the conserved sex chromosomes of snakes, birds, and therian mammals share no orthology by gene content. This nonhomology has generally been attributed to their independent evolution from different autosomes (e.g., Graves 2006); however, such a situation could also arise by independent rearrangements of a large ancestral (sex) chromosome in an amniote ancestor (Ezaz et al. 2006; Smith and Voss 2007).
The latter hypothesis can be tested by comparing the syntenic arrangement of sex chromosome orthologs in amniote outgroups, such as amphibians or fish. Smith and Voss (2007) found that orthologs from chicken 2 (=snake Z), chicken Z, and human X map to the same chromosome (linkage group 2) in the tiger salamander, Ambystoma tigrinum, with which amniotes last shared a common ancestor some 360–390 MYA (citations to these and other divergence dates are made in Fig. 1). However, synteny of chicken and human sex chromosome orthologs is not shared by any other amphibian or amniote so far examined. In A. tigrinum, such an arrangement of genes may be coincidental, made more likely by the very large chromosomes of this species.
Additional outgroup comparison is provided by the recently sequenced genome of the tropical clawed frog, Xenopus tropicalis. The snake Z (chicken chromosomes 2p + 27) is orthologous to parts of Xenopus chromosomes 6 and 10; the chicken Z is orthologous to part of chromosome 1; and the human X is orthologous to parts of chromosomes 2 and 8 (Fig. 1b) (Hellsten et al. 2010; O’Meally et al. 2010); thus, none of these conserved amniote sex chromosomes is syntenic in Xenopus.
Further afield, teleost fish such as Gasterosteus, Danio, Fugu, and Oryzias also provide an appropriate outgroup, but they are not ideal because the teleost genome duplication complicates detection of orthologs (Kasahara et al. 2007) and their genomes are rearranged and gene duplicates have dropped out, so synteny is poorly conserved (Nakatani et al. 2007). However, fine-scale analysis of these genomes produced no evidence of synteny of bird Z and human X orthologs in teleost fish (Bellott et al. 2010).
In their in silico reconstructions of ancestral vertebrate chromosomes, neither Nakatani et al. (2007) nor Kohn et al. (2006) specifically addressed the question of ancestral synteny of amniote sex chromosomes, but their proposed ancestral karyotypes do not recover synteny of snake with bird, or bird with mammal, sex chromosomes. The absence of ancestral synteny in the blocks that make up the sex chromosomes of extant therian mammals, birds, and snakes suggests that they were each derived independently from different ancestral autosomes.
Relationship between fused regions in sex chromosomes
But is there a deeper relationship? In eutherian mammals, two ancestral chromosome segments make up the X. The conserved region (XCR) is defined by the X chromosome in marsupials and the added region (XAR) by an autosomal gene block (Fig. 3) (Graves 1995; Mikkelsen et al. 2007; Ross et al. 2005), and this fused X gave rise to a degenerate Y with a conserved region (YCR) and added region (YAR) (Waters et al. 2001). The same two blocks lie on different autosomes in all other tetrapods for which mapping data are available (Fig. 1b). This suggests that the eutherian X arose by fusion of an ancestral autosome with the therian X between 148 MYA, when marsupials and eutherians last shared a common ancestor and 105 MYA before eutherians radiated. The position of the centromere at this junction on the X of the African elephant, Loxodonta africana, implies that there was a centric fusion in the ancestor of eutherians, followed by a centric shift in the ancestor of other eutherian lineages (Rodríguez Delgado et al. 2009).
The eutherian X chromosome consists of two ancestral chromosome segments known as the X-conserved region (XCR, 45–155 Mbp, blue) which is shared with marsupials, and an added region (XAR, 0–45 Mbp, red) that is autosomal in marsupials. Intriguingly, these two segments derive in large part from a single ancestral vertebrate chromosome, prior to two rounds of genome duplication (Nakatani et al. 2007). Of the 687 human X-borne Ohonologs derived from the jawed-vertebrate whole genome duplication identified by Makino and McLysaght (2010), 20 pairs (green arcs) have a member on both the XAR (red) and XCR (blue), and 2 pairs (magenta arcs) on just the XCR, presumably due to small-scale rearrangements. Perhaps the wholesale addition of Ohnologs allowed the rapid specialization of a “smart and sexy” eutherian X chromosome
Similarly, synteny of genes on the snake Z must have arisen by the fusion of ancestral segments (represented by chicken chromosomes 2p and 27) between 166 and 275 MYA because, with the exception of agamid and iguanid lizards (Srikulnath et al. 2009b, http://ensembl.org release 64), these regions are always found on separate chromosomes in other tetrapods (Fig. 1b).
Were the independent fusions of these segments in mammals and snakes chance events, or were the two ancestral blocks of the X or Z related in some way? One possibility is that they were ancient duplicates that arose through two rounds of vertebrate genome duplication, first proposed by Ohno (1970). By using tunicate and sea urchin genes to define paralogs (“Ohnologs”) that arose through these ancient vertebrate genome duplications, Nakatani et al. (2007) were able to reconstruct proto-karyotypes for the pre-duplicated genome of the ancestral vertebrate; after one round of genome duplication in the ancestral gnathostome (jawed vertebrate); and after two rounds in the ancestral amniote. Inspection of these ancient duplicates implies that a single chromosome in the ancestral vertebrate gave rise to the two segments that would ultimately define the conserved and added regions of the eutherian X (Fig. 3).
The human X chromosome is known to bear an overrepresentation of genes involved in mental retardation (MRX) and reproduction (Graves et al. 2002; Nguyen and Disteche 2006; Saifi and Chandra 1999; Skuse 2005) that is thought to have arisen through gene trafficking and subfunctionalization of transposed copies (Potrzebowski et al. 2008). Alternatively, it might reflect the biased gene content of the ancestral autosomes from which it evolved (Graves 2006). Candidate human MRX genes from the XAR map to a single autosome in marsupials and most other tetrapods, implying that their location on the X arose, not by independent translocations, but in a single addition of the XAR (Delbridge et al. 2008). Some known and candidate MRX genes have widespread expression patterns in chickens and marsupials, but are restricted to neuronal and gonadal tissues or particular cell types in mice and humans (Delbridge et al. 2008; Kohn et al. 2007). The addition of a segment rich in Ohnologs to a chromosome with brain and reproductive functions may have enabled genes on the neo-X of the ancestral eutherian to take on new and highly specialized roles. Combined with the special selective forces that act on X chromosomes (e.g., fast X, sexual selection; Charlesworth et al. 1987; Singh and Petrov 2007), the nascent X was transformed in to a chromosome that is both “smart and sexy” (Graves et al. 2002).
Remarkably, the two segments of the snake Z also derive in large part from two duplicates of a single ancestral vertebrate chromosome (represented by chicken 2q and 27). The Z chromosome appears to be conserved at least across “true” snakes (Alethinophidia) whose origins are in the Cretaceous about 105 MYA; the gene content of sex chromosomes in the earliest snakes (blindsnakes, Scolecophidia), which arose about 160 MYA (Vidal et al. 2009), remains unknown. Gene mapping in lizards indicates that the fusion of these ancestral segments predates the origin of snakes by 6 Ma. Perhaps the distant homology of these segments predisposed subsequent fusion in an early snake ancestor. Such fusions may have been favored by selection because of their complementary gene content. This distant homology may have enabled rapid subfunctionalization of Z-borne genes by selection on the hemizygous Z of females, or sexual selection and fixation of novel mutants arising by drift (Mank et al. 2010). Perhaps this sex chromosome even played some role in the rapid divergence of snakes from other lizards. The fully assembled and mapped genomes of amphioxus (Putnam et al. 2008), several snakes (http://snakegenomics.org), and gene mapping in a diversity of reptiles will inform the interpretation of these observations.
Which came first, the chicken or the Z?
The chicken Z represents a single ancestral chordate chromosome, which is also present as a single chromosome, or chromosome arm, in many reptiles and amphibians (Graves and Shetty 2001; Matsuda et al. 2005; Nakatani et al. 2007; Nanda et al. 2008; Pokorná et al. 2011; Srikulnath et al. 2009b; Voss et al. 2011). In the ancestral karyotypes proposed by Nakatani et al. (2007), synteny of the ancestral vertebrate proto-chromosome “A” is conserved in the gnathostome proto-chromosome “A0,” the amniote proto-chromosomes “3” and “26,” and is found today in chicken chromosomes Z and 17. Almost all orthologs of chicken Z genes map to chromosome 1 in Xenopus (Fig. 1b) (Hellsten et al. 2010) and linkage group 8 in the Mexican axolotl, Ambystoma mexicanum (Voss et al. 2011). Even in the ancestral chordate karyotype suggested by analysis of the amphioxus genome (Putnam et al. 2008), chicken Z orthologs cluster as linkage group 2.
This represents an unusually stable syntenic arrangement, which remained intact for more than 500 Ma, with its origin long before the evolution of birds. It has been proposed that a role in sex determination, and the subsequent accumulation of male-specific genes on the chicken Z chromosome, exerts selective pressure to conserve Z chromosome synteny in birds (Nanda et al. 2008), but clearly synteny has been conserved even in lineages in which it does not act in that role.
The chicken Z and sex determination
The distribution of, and enormous variation in, mechanisms of sex determination across the amniote phylogeny suggests that sex chromosomes have evolved many times (Ezaz et al. 2006; Graves 2008; Organ and Janes 2008; Ezaz et al. 2009b). However, the ancestral chromosome represented by the chicken Z is found to have a sex-determining function in at least three very distantly related lineages. The chicken Z chromosome is shared by all birds, although there are some lineage specific rearrangements (Nanda et al. 2008). Gene mapping in the Hokou gecko, Gekko hokouensis, also reveals a gene content and order in the acrocentric Z and slightly degenerated W that is shared with the chicken Z chromosome (Kawai et al. 2009). Most remarkably, the bizarre platypus sex chromosome system of five Xs and five Ys that form a translocation chain at meiosis share homology with the chicken Z, principally in the terminal X chromosome, X5 (Veyrunes et al. 2008, http://ensembl.org, release 64).
Many reptiles have sex chromosomes that are not homologous to the bird ZW, and their gene content makes it clear that they were independently derived from different autosomes (e.g., Ezaz et al. 2009a; Matsubara et al. 2006; Pokorná et al. 2011; Alfoldi et al. 2011). However, reconstructing ancestral synteny suggests some were derived from the same ancestral chromosome as was the chicken Z, despite the absence of shared synteny in extant species. For instance, the Z chromosome of the Chinese softshell turtle, Pelodiscus sinensis, shares five genes with chicken chromosome 15, and the chicken Z chromosome is homologous to chromosome 6, although the gene order is different (Matsuda et al. 2005). This suggests an origin in a different autosomal pair of chromosomes than the chicken Z (Kawagoshi et al. 2009). However, in opossum and Xenopus these five turtle Z genes (and most other genes in the same region) are syntenic with orthologs from chicken Z (Fig. 1b), suggesting the chromosomes were fused at some stage (Hellsten et al. 2010; Mikkelsen et al. 2007). In amphioxus, scaffolds remain unassigned to chromosomes, but at least one of the five mapped turtle Z genes (TOP3B) lies on a scaffold shared with chicken Z orthologs (Putnam et al. 2008). Strikingly, the X chromosome of the green anole lizard, Anolis carolinensis, also shares synteny with chicken chromosome 15 (Alfoldi et al. 2011). These observations suggest that both the X of Anolis and the Z chromosome of Pelodiscus are derived from the same ancestral autosome that gave rise to the chicken Z. Understanding how this chromosome acts in opposite systems of heterogamety (XY vs. ZW) is of great interest, particularly for comparing fast X and Z effects, or demonstrating differential gene traffic onto or off the Z and X.
Evolution of the sex-determining function of the proto-Z
Homology of sex chromosomes in amniotes as divergent as birds, a gecko and monotreme mammals raises the question of whether the sex-determining function of this 325-Ma-old proto-Z chromosome has been retained in several different descendants, or whether the ancestral chromosome that gave rise to it has an unusual tendency to become a sex chromosome (Graves and Peichel 2010). Testing these alternative hypotheses requires close examination of the sex chromosomes in lineages that contain a bird-like ZW.
Has the sex-determining function of the platypus X5 chromosome been preserved? It has been suggested that the monotreme XY complex evolved by repeated translocations of four autosomes with an ancestral bird-like Z chromosome to form the translocation chain of 10 seen at meiosis (Graves 2008; Rens et al. 2007; Veyrunes et al. 2008). To test this hypothesis, it would be necessary to examine sex determination in related species. However, there are no mammal taxa extant other than Theria, which have recently evolved a unique XY system, so it is not possible to determine whether the monotreme XY system retained an ancestral sex-determining function or gained its sex-determining function independently.
The relationship between the bird ZW and the ZW system of the Hokou gecko, G. hokouensis, is more amenable to analysis. Chromosome painting shows that sex chromosome homology with the chicken Z is not widespread among reptiles (Fig. 4). Pokorná et al. (2011) hybridized a chicken Z chromosome paint to chromosome spreads of species with cytologically identifiable sex chromosomes, representing seven squamate families. In each case, the probe marked only autosomes. A further 18 squamates in 10 families confirm that synteny of the Z is conserved, but sex chromosomes remain unidentified in these species. Of particular interest are species related to G. hokouensis, as they may give an indication of the antiquity of the gecko Z chromosome. Pokorná et al. (2011) examined three species congeneric with G. hokouensis, and found that a single acrocentric chromosome was marked by the chicken Z probe; however, sex chromosomes remain unidentified in these species (Gekko japonicus, Gekko vittatus, and Gekko ulikovskii). Trifonov et al. (2011) used chromosome paints from G. japonicus to demonstrate that sex chromosomes in the tokay gecko, G. gecko, and the flat-tailed house gecko, Hemidactylus platyurus, are not homologous with those in chicken. In more distantly related gekkotans (a pygopod and a eublepharid), XXY sex chromosomes are also not homologous with the chicken Z, suggesting that those of G. hokouensis arose independently and more recently than the sex chromosomes of other gekkotans (Fig. 4) (Pokorná et al. 2011). The alternative explanation that the sex-determining function was lost many times in other squamates and geckoes is less parsimonious. We can conclude, therefore, that the ancestral proto-Z acquired a sex-determining function independently at least twice, once in the ancestors of birds, and once in the ancestors of G. hokouensis.
A simplified phylogeny of geckos (Gekkota), showing chromosomal homology with the chicken Z chromosome (green). Geckos have evolved sex chromosomes independently many times (open chromosomes) and many exhibit temperature-dependent sex determination (bulb). Among all species examined, including other lizards and snakes, a single-chromosome arm is homologous to the chicken Z (with some exceptions). Only in one species, Gekko hokouensis, does the same chromosome have a role in sex determination, suggesting this function arose independently, rather than being retained for ~275 Ma. Mapping and painting data from Kawai et al. (2009), Pokorná et al. (2011), and Trifonov et al. (2011); phylogeny after Gamble et al. (2011) and Rösler et al. (2011)
Does the identity of the sex-determining gene provide a definitive answer on the functional homology of the sex-determining role for the chicken, gecko, and platypus sex chromosomes? In the chicken, the sex-determining gene has been proposed to be DMRT1, which lies on the Z but not the W, and appears to operate by means of the dosage difference between ZZ males and ZW females. Knocking down DMRT1 in genotypically ZZ eggs produces sex-reversed females (Smith et al. 2009). Dosage of DMRT1 is also critical in humans, for deletion of the tip of chromosome 9, where DMRT1 maps in humans, produces XY females (Calvari et al. 2000). It is unknown whether DMRT1 is involved in sex determination in G. hokouensis. In the platypus, it seems unlikely that DMRT1 could play a sex-determining role, given that it is present in one copy in XY males and two copies in XX females (Grützner et al. 2004).
If sex determination were found to be initiated by the same gene(s), would this provide evidence of a shared evolutionary history in sex determination? This would assume no homoplasy, that is, no predisposition to enlist common machinery when new instances of sex determination arise in evolution (Graves and Peichel 2010). This is clearly not a valid assumption, given that DMRT1 has independently spawned novel sex-determining systems in fish and frogs. In medaka, Oryzias latipes, a retroposed copy of DMRT1 acts as a male-dominant testis-determining gene, presumably by boosting the concentration of DMRT1 product (Matsuda et al. 2002). It defines a new Y chromosome that is shared only by the most closely related species. In Xenopus laevis, a partially duplicated copy has the opposite function; lacking a transactivating domain it inhibits DMRT1, so acts as a dominant female-determining “anti-testis” gene (Yoshimoto and Ito 2011). Only the most closely related species share this system, showing that it was recently independently derived.
What could predispose the proto-Z to a sex-determining function?
Comparative maps provide a unique perspective on the evolution of sex chromosomes: different autosomes can be co-opted to this role independently, but some appear to be recruited more frequently than others. What makes some chromosomes more suited to this role than others?
In the case of the chicken Z, perhaps its inclusion of the sex-determining and sex-differentiation gene DMRT1 makes it a particularly good candidate. As discussed above, transposed copies of DMRT1 are thought to be sex determining in medaka and Xenopus, and its two-fold dosage directs male development in the chicken and is necessary in mammals. The primary sex-determining genes are yet to be identified in the Hokou gecko, but having established sex chromosome homology with birds, the list of candidates is narrowed considerably. In monotremes, although DMRT1 is an unlikely candidate, it may have been the original sex-determining gene, which was supplanted by a gene on the autosome with which it first fused.
Perhaps the unusual stability of the proto-Z chromosome also permits it to retain not one, but two, or even several genes that must act in concert to effect sex determination. It is clear that DMRT1 dosage alone cannot explain the occurrence of gynandromorph chickens (Zhao et al. 2010), and there must also be a cell-autonomous factor on one or other sex chromosome to explain the sexually dimorphic features on the ZZ and ZW sides of the same bird (Clinton et al. 2011). The mammal X chromosome, too, is highly stable in gene content and even gene order, though this may be a consequence of selection against disruptions of a chromosome-wide dosage compensation mechanism.
Another possibility is that this proto-Z makes a good candidate for sex chromosomes because the genes it bears are insensitive to haploinsufficiency. This would obviate the need to evolve a complex system of dosage compensation (Livernois et al. 2011). Indeed, dosage compensation is only partial and gene-specific in birds, and also in platypus (Deakin et al. 2008; Itoh et al. 2010). In contrast, failure to inactivate the mammal X chromosome or duplications of even small regions on the active X have severe phenotypes (Migeon et al. 2000; Schmidt and Du Sart 1992).
Others have argued for the role that sexual selection and sexually antagonistic genes play in shaping sex chromosomes. It is possible that autosomes that bear a suite of such genes are preferentially recruited to a role in sex determination (Bachtrog et al. 2011). Pala et al. (2011) report a rare case of autosomal fission and fusion with both Z and W sex chromosomes in warblers and their allies (Sylvioidea). The neo-sex chromosome was formed by fusion of the distal end of the ancestral Z and part of chromosome 4a. This 42-Ma-old rearrangement may have been favored because it brings one or more genes with sex-biased functions (e.g., androgen receptor, AR) into linkage with the male determining gene DMRT1 (Pala et al. 2011).
The huge variety in the mechanisms of sex determination and life history traits among amniotes and, in particular, reptiles, provides countless natural experiments with which to test these and other ideas. Sophisticated molecular cytogenetic techniques and wide taxonomic sampling in genome sequencing (e.g., Genome 10K Community of Scientists 2009) will provide further insights in to the evolutionary history of amniote sex chromosomes.
Abbreviations
- GSD:
-
Genotypic sex determination
- Mbp:
-
Million base pairs
- MRX:
-
Mental retardation genes on the X chromosome
- MYA:
-
Million years ago
- TSD:
-
Temperature-dependent sex determination
- XAR:
-
X added region
- XCR:
-
X conserved region
References
Alfoldi J, Di Palma F, Grabherr M et al (2011) The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591. doi:10.1038/nature10390
Bachtrog D (2006) A dynamic view of sex chromosome evolution. Curr Opin Genet Dev 16:578–585. doi:10.1016/j.gde.2006.10.007
Bachtrog D, Kirkpatrick M, Mank JE et al (2011) Are all sex chromosomes created equal? Trends Genet 27:350–357
Beçak W, Beçak ML, Nazareth HRS, Ohno S (1964) Close karyological kinship between the reptilian suborder Serpentes and the class Aves. Chromosoma 15:606–617
Bellott DW, Page DC (2010) Reconstructing the evolution of vertebrate sex chromosomes. Cold Spring Harb Symp Quant Biol 74:345–353
Bellott DW, Skaletsky H, Pyntikova T et al (2010) Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466:612–616. doi:10.1038/nature09172
Bergero R, Charlesworth D (2009) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24:94–102. doi:10.1016/j.tree.2008.09.010
Bininda-Emonds ORP, Cardillo M, Jones KE et al (2007) The delayed rise of present-day mammals. Nature 446:507–512. doi:10.1038/nature05634
Bull JJ (1983) Evolution of sex determining mechanisms. Benjamin/Cummings Pub. Co., Menlo Park
Calvari V, Bertini V, De Grandi A et al (2000) A new submicroscopic deletion that refines the 9p region for sex reversal. Genomics 65:203–212. doi:10.1006/geno.2000.6160
Charlesworth B, Coyne JA, Barton NH (1987) The relative rates of evolution of sex chromosomes and autosomes. Am Nat 130:113–146
Charnier M (1966) Action de la temperature sur la sex-ratio chez l’embryon d’Agama agama (Agamidae, Lacertilien). Comtes rendus des séances de la societe de biologie et des ses filiales (Paris) 160:620–622
Clinton M, Zhao D, Nandi S and McBride D (2011) Evidence for avian cell autonomous sex identity (CASI) and implications for the sex-determination process?. Chomosoma.doi:10.1007/s10577-011-9257-9
Deakin JE, Hore TA, Koina E, Graves JAM (2008) The status of dosage compensation in the multiple x chromosomes of the platypus. PLoS Genet 4:e1000140. doi:10.1371/journal.pgen.1000140
Delbridge ML, McMillan DA, Doherty RJ et al (2008) Origin and evolution of candidate mental retardation genes on the human X chromosome (MRX). BMC Genom 9:65. doi:10.1186/1471-2164-9-65
Ezaz T, Quinn AE, Miura I et al (2005) The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosom Res 13:763–776
Ezaz T, Stiglec R, Veyrunes F, Graves JAM (2006) Relationships between vertebrate ZW and XY sex chromosome systems. Curr Biol 16:R736–R743. doi:10.1016/j.cub.2006.08.021
Ezaz T, Moritz B, Waters PD et al (2009a) The ZW sex microchromosomes of an Australian dragon lizard share no homology with those of other reptiles or birds. Chromosom Res 17:965–973
Ezaz T, Sarre SD, O’Meally D et al (2009b) Sex chromosome evolution in Lizards: independent origins and rapid transitions. Cytogenet Genome Res 127:249–260. doi:10.1159/000300507
Gamble T (2010) A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex Dev 4:88–103. doi:10.1159/000289578
Gamble T, Bauer AM, Colli GR et al (2011) Coming to America: multiple origins of New World geckos. J Evol Biol 24:231–244. doi:10.1111/j.1420-9101.2010.02184.x
Genome 10K Community of Scientists (2009) Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered 100:659–674. doi:10.1093/jhered/esp086
Georges A, Ezaz T, Quinn AE, Sarre SD (2010) Are reptiles predisposed to temperature-dependent sex determination? Sex Dev 4:7–15
Graves JAM (1995) The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. BioEssays 17:311–320. doi:10.1002/bies.950170407
Graves JAM (2006) Sex chromosome specialization and degeneration in mammals. Cell 124:901–914. doi:10.1016/j.cell.2006.02.024
Graves JAM (2008) Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Genetics 42:565–586. doi:10.1146/annurev.genet.42.110807.091714
Graves JAM, Peichel C (2010) Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol 11:205. doi:10.1186/gb-2010-11-4-205
Graves JAM, Shetty S (2001) Sex from W to Z: evolution of vertebrate sex chromosomes and sex determining genes. J Exp Zool 290:449–462
Graves JAM, Gecz J, Hameister H (2002) Evolution of the human X—a smart and sexy chromosome that controls speciation and development. Cytogenet Genome Res 99:141–145. doi:10.1159/000071585
Grützner F, Rens W, Tsend-Ayush E et al (2004) In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432:913–917. doi:10.1038/nature03021
Hedges SB, Kumar S (2009) The timetree of life. Oxford University Press US
Hellsten U, Harland RM, Gilchrist MJ et al (2010) The genome of the western clawed frog Xenopus tropicalis. Science 328:633–636. doi:10.1126/science.1183670
Hugall AF, Foster R, Lee MSY (2007) Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst Biol 56:543–563. doi:10.1080/10635150701477825
Ito M, Mawaribuchi S, Yoshimoto S, Ohashi S, Takamatsu N (2011) Molecular evolution of vertebrate sex-determining genes. Chromosome Res. doi:10.1007/s10577-011-9265-9
Itoh Y, Replogle K, Kim Y-H et al (2010) Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res 20:512–518. doi:10.1101/gr.102343.109
Kasahara M, Naruse K, Sasaki S et al (2007) The medaka draft genome and insights into vertebrate genome evolution. Nature 447:714–719. doi:10.1038/nature05846
Kawagoshi T, Uno Y, Matsubara K et al (2009) The ZW micro-sex chromosomes of the Chinese soft-shelled turtle (Pelodiscus sinensis, Trionychidae, Testudines) have the same origin as chicken chromosome 15. Cytogenet Genome Res 125:125–131. doi:10.1159/000227837
Kawai A, Nishida-Umehara C, Ishijima J et al (2007) Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet Genome Res 117:92–102. doi:10.1159/000103169
Kawai A, Ishijima J, Nishida C et al (2009) The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118:43–51. doi:10.1007/s00412-008-0176-2
Kohn M, Hogel J, Vogel W et al (2006) Reconstruction of a 450-My-old ancestral vertebrate protokaryotype. Trends Genet 22:203–210. doi:10.1016/j.tig.2006.02.008
Kohn M, Kehrer-Sawatzki H, Steinbach P et al (2007) Recruitment of old genes to new functions: evidences obtained by comparing the orthologues of human XLMR genes in mouse and chicken. Cytogenet Genome Res 116:173–180. doi:10.1159/000098183
Koopman P, Gubbay J, Vivian N et al (1991) Male development of chromosomally female mice transgenic for Sry. Nature 351:117–121. doi:10.1038/351117a0
Livernois AM, Graves JAM, Waters PD (2011) The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity. doi:10.1038/hdy.2011.106
Makino T, McLysaght A (2010) Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A 107:9270–9274. doi:10.1073/pnas.0914697107
Mank JE (2011) Small but mighty: the evolutionary dynamics of W and Y sex chromosomes. Chromosome Res. doi:10.1007/s10577-011-9251-2
Mank JE, Nam K, Ellegren H (2010) Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol 27:661–670. doi:10.1093/molbev/msp282
Matsubara K, Tarui H, Toriba M et al (2006) Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A 103:18190
Matsuda M, Nagahama Y, Shinomiya A et al (2002) DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417:559–563
Matsuda Y, Nishida-Umehara C, Hiroshi T et al (2005) Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosom Res 13:601–615. doi:10.1007/s10577-005-0986-5
Migeon BR, Ausems M, Giltay J et al (2000) Severe phenotypes associated with inactive ring X chromosomes. Am J Med Genet 93:52–57. doi:10.1002/1096-8628(20000703)93:1<52::AID-AJMG9>3.0.CO;2-9
Mikkelsen TS, Wakefield MJ, Aken B et al (2007) Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447:167–177. doi:10.1038/nature05805
Ming R, Gschwend AR , Weingartner LA, Moore RC (2011) The sex-specific region of sex chromosomes in animals and plants. Chromosome Res. doi:10.1007/s10577-011-9255-y
Mittwoch U (2000) Three thousand years of questioning sex determination. Cytogenet Cell Genet 91:186–191. doi:10.1159/000056842
Nakatani Y, Takeda H, Kohara Y, Morishita S (2007) Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res 17:1254–1265. doi:10.1101/gr.6316407
Nanda I, Haaf T, Schartl M et al (2002) Comparative mapping of Z-orthologous genes in vertebrates: implications for the evolution of avian sex chromosomes. Cytogenet Genome Res 99:178–184. doi:10.1159/000071591
Nanda I, Schlegelmilch K, Haaf T et al (2008) Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet Genome Res 122:150–156. doi:10.1159/000163092
Nguyen DK, Disteche CM (2006) High expression of the mammalian X chromosome in brain. Brain Res 1126:46–49. doi:10.1016/j.brainres.2006.08.053
O’Meally D, Miller H, Patel HR et al (2009) The first cytogenetic map of the tuatara, Sphenodon punctatus. Cytogenet Genome Res 127:213–223. doi:10.1159/000300099
O’Meally D, Patel HR, Stiglec R et al (2010) Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. doi:10.1007/s10577-010-9152-9
Ohno S (1967) Sex chromosomes and sex-linked genes. Springer, Germany
Ohno S (1970) Evolution by gene duplication. G. Allen and Unwin, Springer, London
Organ CL, Janes DE (2008) Evolution of sex chromosomes in Sauropsida. Integr Comp Biol 48:512
Ospina-Álvarez N, Piferrer F (2008) Temperature-dependent sex determination in fish revisited: prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS One 3:e2837. doi:10.1371/journal.pone.0002837
Pala I, Naurin S, Stervander M et al (2011) Evidence of a neo-sex chromosome in birds. Heredity. doi:10.1038/hdy.2011.70
Perrin N (2009) Sex reversal: a fountain of youth for sex chromosomes. Evolution 63:3043–3049. doi:10.1111/j.1558-5646.2009.00837.x
Pokorná M, Giovannotti M, Kratochvíl L et al (2011) Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma. doi:10.1007/s00412-011-0322-0
Potrzebowski L, Vinckenbosch N, Marques AC et al (2008) Chromosomal gene movements reflect the recent origin and biology of Therian sex chromosomes. PLoS Biol. doi:10.1371/journal.pbio.0060080
Putnam NH, Butts T, Ferrier DEK et al (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071. doi:10.1038/nature06967
Quinn AE, Georges A, Sarre SD et al (2007) Temperature sex reversal implies sex gene dosage in a reptile. Science 316:411. doi:10.1126/science.1135925
Radder RS, Quinn AE, Georges A et al (2008) Genetic evidence for co-occurrence of chromosomal and thermal sex-determining systems in a lizard. Biol Lett 4:176–178. doi:10.1098/rsbl.2007.0583
Rens W, O’Brien PC, Grutzner F et al (2007) The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol 8:R243. doi:10.1186/gb-2007-8-11-r243
Rhen T, Schroeder A, Sakata J et al (2011) Segregating variation for temperature-dependent sex determination in a lizard. Heredity 106:649–660. doi:10.1038/hdy.2010.102
Rodríguez Delgado C, Waters P, Gilbert C et al (2009) Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosom Res 17:917–926. doi:10.1007/s10577-009-9079-1
Rösler H, Bauer AM, Heinicke MP et al (2011) Phylogeny, taxonomy, and zoogeography of the genus Gekko Laurenti, 1768 with the revalidation of G. reevesii Gray, 1831 (Sauria: Gekkonidae). Zootaxa 2989:1–50
Ross MT, Grafham DV, Coffey AJ et al (2005) The DNA sequence of the human X chromosome. Nature 434:325–337. doi:10.1038/nature03440
Saifi GM, Chandra HS (1999) An apparent excess of sex- and reproduction-related genes on the human X chromosome. Proc Biol Sci 266:203–209
Schmidt M, Du Sart D (1992) Functional disomies of the X chromosome influence the cell selection and hence the X inactivation pattern in females with balanced X–autosome translocations: a review of 122 cases. Am J Med Genet 42:161–169. doi:10.1002/ajmg.1320420205
Shetty S, Griffin DK, Graves JAM (1999) Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosom Res 7:289–295
Sinclair AH, Berta P, Palmer MS et al (1990) A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346:240–244. doi:10.1038/346240a0
Singh N, Petrov D (2007) Evolution of gene function on the X chromosome versus the autosomes. Genome Dynamics 3:101
Skuse DH (2005) X-linked genes and mental functioning. Hum Mol Genet 14:R27–R32. doi:10.1093/hmg/ddi112
Smith JJ, Voss SR (2007) Bird and mammal sex-chromosome orthologs map to the same autosomal region in a salamander (Ambystoma). Genetics 177:607–613. doi:10.1534/genetics.107.072033
Smith CA, Roeszler KN, Ohnesorg T et al (2009) The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461:267–271. doi:10.1038/nature08298
Srikulnath K, Matsubara K, Uno Y et al (2009a) Karyological characterization of the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Squamata) by molecular cytogenetic approach. Cytogenet Genome Res 125:213–223. doi:10.1159/000230005
Srikulnath K, Nishida C, Matsubara K et al (2009b) Karyotypic evolution in squamate reptiles: comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosom Res 17:975–986. doi:10.1007/s10577-009-9101-7
Trifonov VA, Giovannotti M, O’Brien PCM et al (2011) Chromosomal evolution in Gekkonidae. I. Chromosome painting between Gekko and Hemidactylus species reveals phylogenetic relationships within the group. Chromosome Res. doi:10.1007/s10577-011-9241-4
Uno Y, Nishida C, Oshima Y et al (2008) Comparative chromosome mapping of sex-linked genes and identification of sex chromosomal rearrangements in the Japanese wrinkled frog (Rana rugosa, Ranidae) with ZW and XY sex chromosome systems. Chromosome Res 16:637–647. doi:10.1007/s10577-008-1217-7
Veyrunes F, Waters PD, Miethke P et al (2008) Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 18:965–973. doi:10.1101/gr.7101908
Vidal N, Rage J, Coulox A, Hedges SB (2009) Snakes (Serpentes). In: Hedges SB, Kumar S (eds) The Timetree of life. Oxford University Press US, pp 390–397
Voss SR, Kump DK, Putta S et al (2011) Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res 21:1306–1312. doi:10.1101/gr.116491.110
Waters PD, Duffy B, Frost CJ et al (2001) The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet Cell Genet 92:74–79. doi:10.1159/000056872
Werneburg I, Sánchez-Villagra MR (2009) Timing of organogenesis support basal position of turtles in the amniote tree of life. BMC Evol Biol 9:1471–2148
Yoshimoto S, Ito M (2011) A ZZ/ZW–type sex determination in Xenopu laevis. FEBS J 278:1020–1026. doi:10.1111/j.1742-4658.2011.08031.x
Zhao D, McBride D, Nandi S et al (2010) Somatic sex identity is cell autonomous in the chicken. Nature 464:237–242. doi:10.1038/nature08852
Acknowledgments
We are grateful for the comments and suggestions of two anonymous referees, which improved the manuscript plenty. We are also grateful for discussions with and feedback from participants of the Institute for Applied Ecology/Invasive Animals Cooperative Research Centre Science Writers’ Workshop 2011.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: Tariq Ezaz and Jennifer Graves.
Rights and permissions
About this article
Cite this article
O’Meally, D., Ezaz, T., Georges, A. et al. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res 20, 7–19 (2012). https://doi.org/10.1007/s10577-011-9266-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9266-8