Abstract
Background
Global genomic hypomethylation is a common epigenetic event in cancer that mostly results from hypomethylation of repetitive DNA elements. Case–control studies have associated blood leukocyte DNA hypomethylation with several cancers. Because samples in case–control studies are collected after disease development, whether DNA hypomethylation is causal or just associated with cancer development is still unclear.
Methods
In 722 elderly subjects from the Normative Aging Study cohort, we examined whether DNA methylation in repetitive elements (Alu, LINE-1) was associated with cancer incidence (30 new cases, median follow-up: 89 months), prevalence (205 baseline cases), and mortality (28 deaths, median follow-up: 85 months). DNA methylation was measured by bisulfite pyrosequencing.
Results
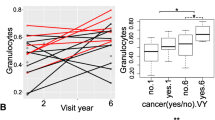
Individuals with low LINE-1 methylation (<median) had a 3.0-fold (95%CI 1.3–6.9) increased incidence of all cancers combined. LINE-1 and Alu methylation were not significantly associated with cancer prevalence at baseline (all cancers combined). However, individuals with low LINE-1 methylation (<median) had a 3.2-fold (95% CI 1.4–7.5) higher prevalence of lung cancer. Individuals with low LINE-1 or Alu methylation (<median) had increased cancer mortality (HR = 3.2, 95%CI 1.3–7.9 for LINE-1; HR = 2.5, 95%CI 1.1–5.8 for Alu).
Conclusion
These findings suggest that individuals with lower repetitive element methylation are at high risk of developing and dying from cancer.
Similar content being viewed by others
References
Ohgane J, Yagi S, Shiota K (2008) Epigenetics: the DNA methylation profile of tissue-dependent and differentially methylated regions in cells. Placenta 29:S29–S35
Lander ES, Linton LM, Birren B et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Wilson AS, Power BE, Molloy PL (2007) DNA hypomethylation and human diseases. Biochim Biophys Acta 1775:138–162
Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32:e38
Gluckman PD, Hanson MA, Cooper C, Thornburg KL (2008) Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 359:61–73
Hsieh CL (2000) Dynamics of DNA methylation pattern. Curr Opin Genet Dev 10:224–228
Das PM, Singal R (2004) DNA methylation and cancer. J Clin Oncol 22:4632–4642
Moore LE, Pfeiffer RM, Poscablo C et al (2008) Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol 9:359–366
Hsiung DT, Marsit CJ, Houseman EA et al (2007) Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 16:108–114
Pufulete M, Al-Ghnaniem R, Leather AJ et al (2003) Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 124:1240–1248
Choi JY, James SR, Link PA et al (2009) Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis 30:1889–1897
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7:11–20
Lelbach A, Muzes G, Feher J (2007) Current perspectives of catabolic mediators of cancer cachexia. Med Sci Monit 13:RA168-173
Gaudet F, Hodgson JG, Eden A et al (2003) Induction of tumors in mice by genomic hypomethylation. Science 300:489–492
Wilson MJ, Shivapurkar N, Poirier LA (1984) Hypomethylation of hepatic nuclear DNA in rats fed with a carcinogenic methyl-deficient diet. Biochem J 218:987–990
Thomas GA, Williams ED (1992) Production of thyroid tumours in mice by demethylating agents. Carcinogenesis 13:1039–1042
Furniss CS, Marsit CJ, Houseman EA, Eddy K, Kelsey KT (2008) Line region hypomethylation is associated with lifestyle and differs by human papillomavirus status in head and neck squamous cell carcinomas. Cancer Epidemiol Biomarkers Prev 17:966–971
Tangkijvanich P, Hourpai N, Rattanatanyong P, Wisedopas N, Mahachai V, Mutirangura A (2007) Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta 379:127–133
Bell B, Rose C, Damon A (1972) The normative aging study: an interdisciplinary and longitudinal study of health and aging. Int J Aging Hum Dev 3:5–17
Bollati V, Baccarelli A, Hou L et al (2007) Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 67:876–880
Tarantini L, Bonzini M, Apostoli P et al (2009) Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect 117:217–222
Jacob RA, Gretz DM, Taylor PC et al (1998) Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr 128:1204–1212
Williams KT, Garrow TA, Schalinske KL (2008) Type I diabetes leads to tissue-specific DNA hypomethylation in male rats. J Nutr 138:2064–2069
Baccarelli A, Wright RO, Bollati V et al (2009) Low blood DNA methylation determines higher risk and mortality from ischemic heart disease and stroke among elderly individuals. Circulation 119:E281
Lim U, Flood A, Choi SW et al (2008) Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology 134:47–55
Bollati V, Schwartz J, Wright R et al (2009) Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 130:234–239
Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD (2009) Prenatal Tobacco Smoke Exposure Affects Global and Gene-Specific DNA Methylation. Am J Respir Crit Care Med 180:462–467
Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC (2008) Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect 116:1547–1552
Hillemacher T, Frieling H, Moskau S et al (2008) Global DNA methylation is influenced by smoking behaviour. Eur Neuropsychopharmacol 18:295–298
Bjornsson HT, Sigurdsson MI, Fallin MD et al (2008) Intra-individual change over time in DNA methylation with familial clustering. JAMA 299:2877–2883
Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7:21–33
Steinhoff C, Schulz WA (2003) Transcriptional regulation of the human LINE-1 retrotransposon L1.2B. Mol Genet Genomics 270:394–402
Babushok DV, Kazazian HH Jr (2007) Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat 28:527–539
Lin CH, Hsieh SY, Sheen IS et al (2001) Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res 61:4238–4243
Soares J, Pinto AE, Cunha CV et al (1999) Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer 85:112–118
Smith IM, Mydlarz WK, Mithani SK, Califano JA (2007) DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer 121:1724–1728
Ogino S, Nosho K, Kirkner GJ et al (2008) A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 100:1734–1738
Cho NY, Kim BH, Choi M et al (2007) Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol 211:269–277
Widschwendter M, Apostolidou S, Raum E et al (2008) Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS One 3:e2656
Zhu ZZ, Hou L, Bollati V et al (2010) Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol doi:10.1093/ije/dyq154
Terry MB, Ferris JS, Pilsner R et al (2008) Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev 17:2306–2310
El-Maarri O, Becker T, Junen J et al (2007) Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet 122:505–514
Choi IS, Estecio MR, Nagano Y et al (2007) Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors). Mod Pathol 20:802–810
Li TH, Schmid CW (2001) Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene 276:135–141
Hou L, Wang H, Sartori S et al (2010) Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer 127:1866–1874
Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3:415–428
von Sternberg RM, Novick GE, Gao GP, Herrera RJ (1992) Genome canalization: the coevolution of transposable and interspersed repetitive elements with single copy DNA. Genetica 86:215–246
Biemont C, Vieira C (2006) Genetics: junk DNA as an evolutionary force. Nature 443:521–524
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences grants ES015172-01; New Investigator funding from the HSPH-NIEHS Center for Environmental Health (ES000002); Environmental Protection Agency grants R83241601 and R827353; and Italian Association for Cancer Research (AIRC-6016). The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, ZZ., Sparrow, D., Hou, L. et al. Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence, and mortality in elderly individuals: the Normative Aging Study. Cancer Causes Control 22, 437–447 (2011). https://doi.org/10.1007/s10552-010-9715-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10552-010-9715-2