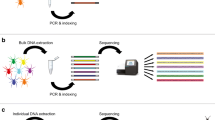
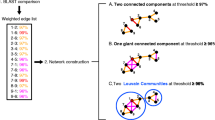
Abstract
The network theoretical framework of ecological community studies is expected to promote not only the basic understanding of ecological and coevolutionary dynamics but also the application of those scientific insights into ecosystem management. However, our knowledge of ecological network architecture in the wild largely stems from empirical studies on macro-organismal systems such as those of plant–pollinator, plant–seed disperser, and prey–predator interactions. In this sense, we have remained ignorant of the diversity of ecological network architecture, its underlying assembly processes, and its consequences on ecological and coevolutionary dynamics. In this paper, I discuss how the high-throughput DNA barcoding of microbes, especially that based on next-generation sequencing, potentially expands the target of ecological network studies. I review the methodological platforms of next-generation sequencing-based analyses of microbe–host animal/plant networks and then introduce some case studies on the networks of plants and their hyper-diverse fungal symbionts. As those preliminary studies are uncovering the unexpected diversity of ecological network architecture, further application of such next-generation sequencing-based analyses to a diverse array of microbial systems will significantly improve our views on community ecological and coevolutionary processes.




Similar content being viewed by others
References
Abarenkov K, Nilsson RH, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vralstad T, Liimatainen K, Peintner U, Koljalg U (2010) The UNITE database for molecular identification of fungi: recent updates and future perspectives. New Phytol 186:281–285
Allen MF (1991) The ecology of mycorrhizae. Cambridge University Press, Cambridge
Allesina S, Tang S (2012) Stability criteria for complex ecosystems. Nature 483:205–208
Almeida-Neto M, Ulrich W (2011) A straightforward computational approach for measuring nestedness using quantitative matrices. Env Model Softw 26:173–178
Almeida-Neto M, Guimarães PR Jr, Lewinsohn TM (2007) On nestedness analyses: rethinking matrix temperature and anti-nestedness. Oikos 116:716–722
Almeida-Neto M, Guimarães P, Guimarães PR Jr, Loyola RD, Ulrich W (2008) A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos 117:1227–1239
Bahram M, Harend H, Tedersoo L (2013) Network perspectives of ectomycorrhizal associations. Fungal Ecol 7:70–77
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376
Ball SL, Armstrong KF (2008) Rapid, one-step DNA extraction for insect pest identification by using DNA barcodes. J Econom Entomol 101:523–532
Barber MJ (2007) Modularity and community detection in bipartite networks. Phys Rev E 76:066102
Bascompte J, Jordano P (2014) Mutualistic networks. Princeton University Press, Princeton
Bascompte J, Jordano P, Melián CJ, Olesen JM (2003) The nested assembly of plant-animal mutualistic networks. Proc Natl Acad Sci USA 100:9383–9387
Bastolla U, Fortuna MA, Pascual-Garía A, Ferrera A, Luque B, Bascompte J (2009) The architecture of mutualistic networks minimizes competition and increases biodiversity. Nature 458:1018–1020
Beiler KJ, Durall DM, Simard SW, Maxwell SA, Kretzer AM (2009) Architecture of the wood-wide web: Rhizopogon spp. genets link multiple Douglas-fir cohorts. New Phytol 185:543–553
Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H (2010) ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189
Bever JD (2003) Soil community feedback and the coexistence of competitors: conceptual frameworks and empirical tests. New Phytol 157:465–473
Blüthgen N, Menzel F, Blüthgen N (2006) Measuring specialization in species interaction networks. BMC Ecol 6:9
Blüthgen N, Menzel F, Hovestadt T, Fiala B (2007) Specialization, constraints, and conflicting interests in mutualistic networks. Curr Biol 17:341–346
Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, Yu DW, de Bruyn M (2014) Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 29:358–367
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Caruso T, Rillig MC, Garlaschelli D (2012) On the application of network theory to arbuscular mycorrhizal fungi–plant interactions: the importance of basic assumptions. New Phytol 194:891–894
CBOL Plant Working Group (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106:12794–12797
Chagnon P, Bradley R, Klironomos J (2014) Plant–fungal symbioses as ecological networks: the need to characterize more than just interaction patterns. Fungal Ecol. doi:10.1016/j.funeco.2014.05.002
Chen S, Yao H, Han J, Liu C, Song J, Shi L, Zhu Y, Ma X, Gao T, Pang X (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5:e8613
China Plant BOL Group (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA 108:19641–19646
Clauset A, Moore C, Newman ME (2008) Hierarchical structure and the prediction of missing links in networks. Nature 453:98–101
Coleman AW (2003) ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genetics 19:370–375
Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA (2012) The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262
Darwin C (1862) On the various contrivances by which British and foreign orchids are fertilized by insects. Murray, London
de Cárcer DA, Denman SE, McSweeney C, Morrison M (2011) Evaluation of subsampling-based normalization strategies for tagged high-throughput sequencing data sets from gut microbiomes. Appl Env Microbiol 77:8795–8798
Donatti CI, Guimarães PR Jr, Galetti M, Pizo MA, Marquitti F, Dirzo R (2011) Analysis of a hyper-diverse seed dispersal network: modularity and underlying mechanisms. Ecol Lett 14:773–781
Dormann CF, Fründ J, Blüthgen N, Gruber B (2009) Indices, graphs and null models: analyzing bipartite ecological networks. Open Ecol J 2:7–24
Frey KG, Herrera-Galeano JE, Redden CL, Luu TV, Servetas SL, Mateczun AJ, Mokashi VP, Bishop-Lilly KA (2014) Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood. BMC Genom 15:96
Fukami T, Nakajima M (2013) Complex plant–soil interactions enhance plant species diversity by delaying community convergence. J Ecol 101:316–324
Fukatsu T, Tsuchida T, Nikoh N, Koga R (2001) Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl Env Microbiol 67:1284–1291
Guimarães PR Jr, Raimundo RLG, Cagnolo L, Melián CJ, Vázquez DP, Goldberg J, Naik R (2012) Interaction Web DataBase. http://www.nceas.ucsb.edu/interactionweb/index.html
Guimarães PR Jr, Jordano P, Thompson JN (2011) Evolution and coevolution in mutualistic networks. Ecol Lett 14:877–885
Guimerà R, Amaral LAN (2005) Cartography of complex networks: modules and universal roles. J Stat Mech Theor Exper 2005:P02001
Hajibabaei M, Singer GA, Hebert PD, Hickey DA (2007) DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genetics 23:167–172
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R (2008) Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5:235–237
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685
Hebert PD, Cywinska A, Ball SL, de Waard JR (2003a) Biological identifications through DNA barcodes. Proc R Soc B Lond 270:313–321
Hebert PD, Ratnasingham S, de Waard JR (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc B Lond 270:S96–S99
Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA 101:14812–14817
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH (2011) Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biol Rev 25:38–47
Hongoh Y, Ekpornprasit L, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Noparatnaraporn N, Kudo T (2006) Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol Ecol 15:505–516
Human Microbiome Project Consortium (2012a) A framework for human microbiome research. Nature 486:215–221
Human Microbiome Project Consortium (2012b) Structure, function and diversity of the healthy human microbiome. Nature 486:207–214
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Hutchinson GE (1961) The paradox of the plankton. Am Nat 95:137–145
Joly S, Davies TJ, Archambault A, Bruneau A, Derry A, Kembel SW, Peres-Neto P, Vamosi J, Wheeler TA (2014) Ecology in the age of DNA barcoding: the resource, the promise and the challenges ahead. Mol Ecol Res 14:221–232
Kadowaki K, Sato H, Yamamoto S, Tanabe AS, Hidaka A, Toju H (2014) Detection of the horizontal spatial structure of soil fungal communities in a natural forest. Popul Ecol 56:301–310
Kato M, Kakutani T, Inoue T, Itino T (1990) Insect–flower relationship in the primary beech forest of Ashu, Kyoto: an overview of the flowering phenology and the seasonal pattern of insect visits. Contr Biol Lab Kyoto Univ 27:309–376
Kiers ET, Denison RF (2008) Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Ann Rev Ecol Evol Syst 39:215–236
Kiers ET, Rousseau RA, West SA, Denison RF (2003) Host sanctions and the legume-rhizobium mutualism. Nature 425:78–81
Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, Palmaer MT, West SA, Vandenkoornhuyse P, Jansa J, Bücking H (2011) Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333:880–882
Kikuchi Y (2008) Endosymbiotic bacteria in insects: their diversity and culturability. Microb Env 24:195–204
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1
Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417:67–70
Kondoh M (2008) Building trophic modules into a persistent food web. Proc Natl Acad Sci USA 105:16631–16635
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102:8369–8374
Lian C, Narimatsu M, Nara K, Hogetsu T (2006) Tricholoma matsutake in a natural Pinus densiflora forest: correspondence between above- and below-ground genets, association with multiple host trees and alteration of existing ectomycorrhizal communities. New Phytol 171:825–836
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J (2013) Fungal community analysis by high-throughput sequencing of amplified markers: a user’s guide. New Phytol 199:288–299
Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M (2012) Comparison of next-generation sequencing systems. J Biomed Biotech 201:251364. doi:10.1155/2012/251364
Mangan SA, Schnitzer SA, Herre EA, Mack KML, Valencia MC, Sanchez EI, Bever JD (2010) Negative plant–soil feedback predicts tree–species relative abundance in a tropical forest. Nature 466:752–755
Mardis ER (2013) Next-generation sequencing platforms. Ann Rev Analyt Chem 6:287–303
Marquitti FMD, Guimarães PR Jr, Pires MM, Bittencourt LF (2013) MODULAR: Software for the autonomous computation of modularity in large network sets. Ecography 37:221–224
May RM (1972) Will a large complex system be stable? Nature 238:413–414
May RM (2006) Network structure and the biology of populations. Trends Ecol Evol 21:394–399
Minamoto T, Yamanaka H, Takahara T, Honjo MN, Kawabata ZI (2012) Surveillance of fish species composition using environmental DNA. Limnology 13:193–197
Montesinos-Navarro A, Segarra-Moragues JG, Valiente-Banuet A, Verdú M (2012) The network structure of plant-arbuscular mycorrhizal fungi. New Phytol 194:536–547
Moran NA (2007) Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA 104:8627–8633
Mougi A, Kondoh M (2012) Diversity of interaction types and ecological community stability. Science 337:349–351
Newman ME, Girvan M (2004) Finding and evaluating community structure in networks. Phys Rev E 69:026113
Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH (2008) Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinfo 4:193–201
Nilsson HR, Tedersoo L, Lindahl BD, Kjøller R, Carlsen T, Quince C, Abarenkov K, Pennanen T, Stenlid J, Bruns T (2011) Towards standardization of the description and publication of next-generation sequencing datasets of fungal communities. New Phytol 191:314–318
Oksanen J, Blanachet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2012) Vegan: community ecology package. R package version 2.0-3. http://cran.r-project.org/web/packages/vegan/index.html
Olesen JM, Bascompte J, Dupont YL, Jordano P (2007) The modularity of pollination networks. Proc Natl Acad Sci USA 104:19891–19896
Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA (1986) Microbial ecology and evolution: a ribosomal RNA approach. Ann Rev Microbiol 40:337–365
Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser SS, Cepicka I, Decelle J, Dunthorn M (2012) CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol 10:e1001419
Peay KG, Kennedy PG, Bruns TD (2008) Fungal community ecology: a hybrid beast with a molecular master. Bioscience 58:799–810
Pimm SL (1991) The balance of nature? Ecological issues in the conservation of species and communities. University of Chicago Press, Chicago
Poisot T, Péquin B, Gravel D (2013) High-throughput sequencing: a roadmap toward community ecology. Ecol Evol 3:1125–1139
Polis GA, Myers CA, Holt RD (1989) The ecology and evolution of intraguild predation: potential competitors that eat each other. Ann Rev Ecol Syst 20:297–330
Pompanon F, Deagle BE, Symondson WO, Brown DS, Jarman SN, Taberlet P (2012) Who is eating what: diet assessment using next generation sequencing. Mol Ecol 21:1931–1950
Prosser JI, Bohannan BJ, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ (2007) The role of ecological theory in microbial ecology. Nat Rev Microbiol 5:384–392
Randall JE (1967) Food habits of reef fishes of the West Indies. Stud Trop Oceanogr 5:665–847
Rappé MS, Giovannoni SJ (2003) The uncultured microbial majority. Ann Rev Microbiol 57:369–394
Rezende EL, Lavabre JE, Guimarães PR Jr, Jordano P, Bascompte J (2007) Non-random coextinctions in phylogenetically structured mutualistic networks. Nature 448:925–928
Ratnasingham S, Hebert PD (2007) BOLD: The barcode of life data system (http://www.barcodinglife.org). Mol Ecol Notes 7:355–364
Robertson C (1928) Flowers and insects lists of visitors of four hundred and fifty three flowers. The Science Press, Lancaster
Rohr RP, Saavedra S, Bascompte J (2014) On the structural stability of mutualistic systems. Science 345(6195):1253497. doi:10.1126/science.1253497
Schleuning M, Ingmann L, Strauß R, Fritz SA, Dalsgaard B, Matthias Dehling D, Plein M, Saavedra F, Sandel B, Svenning JC (2014) Ecological, historical and evolutionary determinants of modularity in weighted seed-dispersal networks. Ecol Lett 17:454–463
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Env Microbiol 75:7537–7541
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246
Seifert KA (2009) Progress towards DNA barcoding of fungi. Mol Ecol Res 9:83–89
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Elsevier, New York
Sommer DD, Delcher AL, Salzberg SL, Pop M (2007) Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64
Staniczenko PP, Kopp JC, Allesina S (2013) The ghost of nestedness in ecological networks. Nat Commun 4:1391
Sullivan DJ, Völkl W (1999) Hyperparasitism: multitrophic ecology and behavior. Ann Rev Entomol 44:291–315
Suweis S, Simini F, Banavar JR, Maritan A (2013) Emergence of structural and dynamical properties of ecological mutualistic networks. Nature 500:449–452
Tanabe AS (2012) Assams, a software distributed by the author at http://www.fifthdimension.jp/
Tanabe AS, Toju H (2013) Two new computational methods for universal DNA barcoding: A benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS One 8:e76910
Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Bahram M, Bechem E, Chuyong G, Kõljalg U (2010) 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188:291–301
Thébault E, Fontaine C (2010) Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329:853–856
Thomas T, Gilbert J, Meyer F (2012) Metagenomics-a guide from sampling to data analysis. Microb Inform Exp 2:3
Thompson JN (2005) The geographic mosaic of coevolution. The University of Chicago Press, Chicago
Thompson JN (2006) Mutualistic webs of species. Science 312:372–373
Toju H, Fukatsu T (2011) Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol Ecol 20:853–868
Toju H, Tanabe AS, Yamamoto S, Sato H (2012) High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7:e40863
Toju H, Yamamoto S, Sato H, Tanabe AS, Gilbert GS, Kadowaki K (2013a) Community composition of root-associated fungi in a Quercus-dominated temperate forest: “codominance” of mycorrhizal and root-endophytic fungi. Ecol Evol 3:1281–1293
Toju H, Sato H, Yamamoto S, Kadowaki K, Tanabe AS, Yazawa S, Nishimura O, Agata K (2013b) How are plant and fungal communities linked to each other in belowground ecosystems? A massively parallel pyrosequencing analysis of the association specificity of root-associated fungi and their host plants. Ecol Evol 3:3112–3124
Toju H, Yamamoto S, Sato H, Tanabe AS (2013c) Sharing of diverse mycorrhizal and root-endophytic fungi among plant species in an oak-dominated cool–temperate forest. PLoS ONE 8:e78248
Toju H, Sato H, Tanabe AS (2014a) Diversity and spatial structure of belowground plant–fungal symbiosis in a mixed subtropical forest of ectomycorrhizal and arbuscular mycorrhizal plants. PLoS One 9:e86566
Toju H, Guimarães PR Jr, Olesen JM, Thompson JN (2014b) Assembly of complex plant–fungus networks. Nature Commun 5:5273
Tringe SG, Von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC (2005) Comparative metagenomics of microbial communities. Science 308:554–557
Tylianakis JM, Tscharntke T, Lewis OT (2007) Habitat modification alters the structure of tropical host-parasitoid food webs. Nature 445:202–205
Tylianakis JM, Didham RK, Bascompte J, Wardle DA (2008) Global change and species interactions in terrestrial ecosystems. Ecol Lett 11:1351–1363
Tylianakis JM, Lalibert E, Nielsen A, Bascompte J (2009) Conservation of species interaction networks. Biol Cons 143:2270–2279
Valentini A, Pompanon F, Taberlet P (2009) DNA barcoding for ecologists. Trends Ecol Evol 24:110–117
van der Putten WH, Bardgett RD, Bever JD, Bezemer TM, Casper BB, Fukami T, Kardol P, Klironomos JN, Kulmatiski A, Schweitzer JA (2013) Plant–soil feedbacks: the past, the present and future challenges. J Ecol 101:265–276
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Wardle DA (2002) Communities and ecosystems: linking the aboveground and belowground components. Princeton University Press, Princeton
Wirta HK, Hebert PD, Kaartinen R, Prosser SW, Várkonyi G, Roslin T (2014) Complementary molecular information changes our perception of food web structure. Proc Natl Acad Sci USA 111:1885–1890
Yamamoto S, Sato H, Tanabe AS, Hidaka A, Kadowaki K, Toju H (2014) Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLoS One 9:e96363
Acknowledgments
I thank Akifumi S. Tanabe for his advice on bioinformatic pipelines and three anonymous reviewers for their constructive comments on the manuscript. This study was supported by the Funding Program for Next Generation World-Leading Researchers of Cabinet Office, the Japanese Government (GS014) and JSPS KAKENHI (No. 26711026).
Conflict of interest
The author declares no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
This manuscript was submitted for the special feature based on a symposium in Osaka, Japan, held on 12 October 2013.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Toju, H. High-throughput DNA barcoding for ecological network studies. Popul Ecol 57, 37–51 (2015). https://doi.org/10.1007/s10144-014-0472-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10144-014-0472-z