Abstract
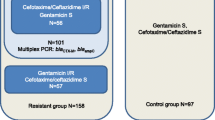
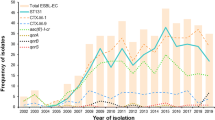
Rapid bacterial typing is a valuable and necessary tool in the prevention and detection of outbreaks. The purpose of this study was to adapt a multilocus variable number of tandem repeats analysis (MLVA) for analysis on a benchtop capillary electrophoresis instrument and compare the modified assay with multilocus sequence typing (MLST) for typing cefpodoxime-resistant Escherichia coli (E. coli). Further, we identified the causative resistance mechanisms and epidemiological type of infection for isolates producing extended-spectrum β-lactamases (ESBLs). A collection of E. coli resistant to cefpodoxime was typed by MLST and a modified MLVA assay using a benchtop capillary electrophoresis instrument. Resistance mechanisms were identified by polymerase chain reaction (PCR) and sequencing. Patient history was examined to establish the epidemiological type of infection for ESBL-producing E. coli. MLVA yielded typing results homologous with MLST and it correctly identified E. coli sequence type (ST) 131 that was accounting for 45 % of all ESBL-producing isolates in the sample collection. The majority (76.7 %) of ESBL-producing isolates was healthcare-related and only 23.3 % of the ESBL-producing isolates were community-onset infections (COI), regardless of the ST. Patients with COI were significantly more often of female gender and younger age compared to healthcare-associated infections (HCAI) and hospital-onset infections (HOI). In conclusion, the modified MLVA is a useful tool for the rapid typing of E. coli and it identified ST131 as the predominating ESBL-producing lineage in Copenhagen. Healthcare-related infections were the predominant infection setting of ESBL-producing E. coli and the demographic characteristics differed between patients with COI and healthcare-related infections.
Similar content being viewed by others
References
Maiden MCJ (2006) Multilocus sequence typing of bacteria. Annu Rev Microbiol 60:561–588
van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M; European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM) (2007) Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13(Suppl 3):1–46
Keys C, Kemper S, Keim P (2005) Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J Appl Microbiol 98:928–940
Noller AC, McEllistrem MC, Pacheco AG, Boxrud DJ, Harrison LH (2003) Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J Clin Microbiol 41:5389–5397
Manges AR, Tellis PA, Vincent C, Lifeso K, Geneau G, Reid-Smith RJ, Boerlin P (2009) Multi-locus variable number tandem repeat analysis for Escherichia coli causing extraintestinal infections. J Microbiol Methods 79:211–213
Lindstedt BA, Brandal LT, Aas L, Vardund T, Kapperud G (2007) Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J Microbiol Methods 69:197–205
Pitout JD, Laupland KB (2008) Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166
Pitout JD, Gregson DB, Campbell L, Laupland KB (2009) Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob Agents Chemother 53:2846–2851
Rooney PJ, O’Leary MC, Loughrey AC, McCalmont M, Smyth B, Donaghy P, Badri M, Woodford N, Karisik E, Livermore DM (2009) Nursing homes as a reservoir of extended-spectrum beta-lactamase (ESBL)-producing ciprofloxacin-resistant Escherichia coli. J Antimicrob Chemother 64:635–641
Rogers BA, Sidjabat HE, Paterson DL (2011) Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14
Van der Bij AK, Peirano G, Pitondo-Silva A, Pitout JD (2012) The presence of genes encoding for different virulence factors in clonally related Escherichia coli that produce CTX-Ms. Diagn Microbiol Infect Dis 72:297–302
Peirano G, Richardson D, Nigrin J, McGeer A, Loo V, Toye B, Alfa M, Pienaar C, Kibsey P, Pitout JD (2010) High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-beta-lactamase-producing Escherichia coli isolates from Canada. Antimicrob Agents Chemother 54:1327–1330
Hansen DS, Frimodt-Møller N; DANRES Study Group (2009) Extended spectrum beta-lactamase producing bacteria in Danish pigs, Danish and imported retail meat and human patients. DANMAP 2009, pp. 19–21. Available online at: http://www.danmap.org/Downloads/Reports.aspx
Nielsen JB, Skov MN, Jørgensen RL, Heltberg O, Hansen DS, Schønning K (2011) Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur J Clin Microbiol Infect Dis 30:773–778
Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL, Reller LB, Sexton DJ (2002) Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 137:791–797
Jørgensen RL, Nielsen JB, Friis-Møller A, Fjeldsøe-Nielsen H, Schønning K (2010) Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J Antimicrob Chemother 65:460–464
Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151
Løbersli I, Haugum K, Lindstedt BA (2012) Rapid and high resolution genotyping of all Escherichia coli serotypes using 10 genomic repeat-containing loci. J Microbiol Methods 88:134–139
Novais A, Pires J, Ferreira H, Costa L, Montenegro C, Vuotto C, Donelli G, Coque TM, Peixe L (2012) Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother 56:3973–3976
Lau SH, Kaufmann ME, Livermore DM, Woodford N, Willshaw GA, Cheasty T, Stamper K, Reddy S, Cheesbrough J, Bolton FJ, Fox AJ, Upton M (2008) UK epidemic Escherichia coli strains A–E, with CTX-M-15 beta-lactamase, all belong to the international O25:H4-ST131 clone. J Antimicrob Chemother 62:1241–1244
Severin JA, Mertaniasih NM, Kuntaman K, Lestari ES, Purwanta M, Lemmens-Den Toom N, Duerink DO, Hadi U, van Belkum A, Verbrugh HA, Goessens WH; Study Group ‘Antimicrobial Resistance in Indonesia: Prevalence and Prevention’ (AMRIN) (2010) Molecular characterization of extended-spectrum beta-lactamases in clinical Escherichia coli and Klebsiella pneumoniae isolates from Surabaya, Indonesia. J Antimicrob Chemother 65:465–469
Kiratisin P, Apisarnthanarak A, Laesripa C, Saifon P (2008) Molecular characterization and epidemiology of extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother 52:2818–2824
Önnberg A, Mölling P, Zimmermann J, Söderquist B (2011) Molecular and phenotypic characterization of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases with focus on CTX-M in a low-endemic area in Sweden. APMIS 119:287–295
Lascols C, Hackel M, Hujer AM, Marshall SH, Bouchillon SK, Hoban DJ, Hawser SP, Badal RE, Bonomo RA (2012) Using nucleic acid microarrays to perform molecular epidemiology and detect novel β-lactamases: a snapshot of extended-spectrum β-lactamases throughout the world. J Clin Microbiol 50:1632–1639
Ben-Ami R, Rodríguez-Baño J, Arslan H, Pitout JD, Quentin C, Calbo ES, Azap OK, Arpin C, Pascual A, Livermore DM, Garau J, Carmeli Y (2009) A multinational survey of risk factors for infection with extended-spectrum beta-lactamase-producing enterobacteriaceae in nonhospitalized patients. Clin Infect Dis 49:682–690
Rodríguez-Baño J, Picón E, Gijón P, Hernández JR, Cisneros JM, Peña C, Almela M, Almirante B, Grill F, Colomina J, Molinos S, Oliver A, Fernández-Mazarrasa C, Navarro G, Coloma A, López-Cerero L, Pascual A (2010) Risk factors and prognosis of nosocomial bloodstream infections caused by extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol 48:1726–1731
Cornejo-Juárez P, Pérez-Jiménez C, Silva-Sánchez J, Velázquez-Acosta C, González-Lara F, Reyna-Flores F, Sánchez-Pérez A, Volkow-Fernández P (2012) Molecular analysis and risk factors for Escherichia coli producing extended-spectrum β-lactamase bloodstream infection in hematological malignancies. PLoS One 7:e35780
Freeman JT, McBride SJ, Heffernan H, Bathgate T, Pope C, Ellis-Pegler RB (2008) Community-onset genitourinary tract infection due to CTX-M-15-producing Escherichia coli among travelers to the Indian subcontinent in New Zealand. Clin Infect Dis 47:689–692
Tängdén T, Cars O, Melhus A, Löwdin E (2010) Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother 54:3564–3568
Wickramasinghe NH, Xu L, Eustace A, Shabir S, Saluja T, Hawkey PM (2012) High community faecal carriage rates of CTX-M ESBL-producing Escherichia coli in a specific population group in Birmingham, UK. J Antimicrob Chemother 67:1108–1113
Hawser SP, Bouchillon SK, Hoban DJ, Badal RE, Hsueh PR, Paterson DL (2009) Emergence of high levels of extended-spectrum-beta-lactamase-producing gram-negative bacilli in the Asia-Pacific region: data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) program, 2007. Antimicrob Agents Chemother 53:3280–3284
Acknowledgment
This work was supported by Fondation Idella (3.3.5/2008/II to K.S.), Fonden til Lægevidenskabens Fremme (10–316 to K.S.), and Hvidovre Hospitals Forskningsfond (grant to J.B.N.).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
(PDF 39 kb)
Rights and permissions
About this article
Cite this article
Nielsen, J.B., Albayati, A., Jørgensen, R.L. et al. An abbreviated MLVA identifies Escherichia coli ST131 as the major extended-spectrum β-lactamase-producing lineage in the Copenhagen area. Eur J Clin Microbiol Infect Dis 32, 431–436 (2013). https://doi.org/10.1007/s10096-012-1764-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-012-1764-x