Abstract
In order to reconstruct the history of Y-chromosome haplogroup (hg) N dispersals in north Eurasia, we have analyzed the diversity of microsatellite (STR) loci within two major hg N clades, N2 and N3, in a total sample of 1,438 males from 17 ethnic groups, mainly of Siberian and Eastern European origin. Based on STR variance analysis we observed that hg N3a is more diverse in Eastern Europe than in south Siberia. However, analysis of median networks showed that there are two STR subclusters of hg N3a, N3a1 and N3a2, that are characterized by different genetic histories. Age calculation of STR variation within subcluster N3a1 indicated that its first expansion occurred in south Siberia [approximately 10,000 years (ky)] and then this subcluster spread into Eastern Europe where its age is around 8 ky ago. Meanwhile, younger subcluster N3a2 originated in south Siberia (probably in the Baikal region) approximately 4 ky ago. Median network and variance analyses of STR haplotypes suggest that south Siberian N3a2 haplotypes spread further into Volga-Ural region undergoing serial bottlenecks. In addition, median network analysis of STR data demonstrates that haplogroup N2-A is represented by two subclusters, showing recent expansion times. The data obtained allow us to suggest Siberian origin of haplogroups N3 and N2 that are currently widespread in some populations of Eastern Europe.
Similar content being viewed by others
Introduction
Molecular phylogeography of Y-chromosome haplogroups provides intriguing clues into the history of human populations, especially in reconstructing of ancient migrations and more recent gene flow episodes (Underhill 2003). One of the most widespread Y-haplogroups in north Eurasia is haplogroup N defined by markers M231 and LLY22g (Jobling and Tyler-Smith 1995; Cinnioglu et al. 2004). This haplogroup is represented by subclades N*, N1, N2 and N3, which are thought to be historically informative in north Eurasian populations (Rootsi et al. 2007). It has been generally accepted that the hg N ancestor is of east Asian ancestry, because haplotypes belonging to ancient paragroup N* are still widely distributed in southeastern Asia and, rarely, even in south Siberia (Rootsi et al. 2007; Sengupta et al. 2006). However, the origin and migration ways of hgs N2 and N3 are not fully understood. According to the first studies, the most likely place of origin of these Y-chromosomes is the area of Mongolia and north China (Zerjal et al. 1997; Karafet et al. 2002). However, substantial frequency of hgs N2 and N3 in Eastern Europe, mainly in Finno-Ugric and Turkic-speaking populations, indicates that these haplogroups may have originated in Eastern Europe (Rosser et al. 2000; Underhill et al. 2001). Moreover, STR diversity within hg N3 has been detected on higher level in Eastern Europeans than in Siberians, which can be interpreted as supporting an origin of hg N3a in Eastern Europe (Villems et al. 2002). Recently, phylogeographic study of hg N structure in north Eurasian populations has demonstrated that south Siberia could be a place of transition of hg N (including N3a and N2) westward to Eastern Europe (Rootsi et al. 2007). However, there is no evidence still that the age of accumulated STR variation within hg N3a is higher in south Siberia.
In this study, we improve our understanding of hg N phylogeographic structure by examining STR variation in a large number of individuals of south Siberian and Eastern European origin. The results obtained give a clear evidence of that hg N3a is older in south Siberia than in Eastern Europe. In addition, the substructure of hg N2-A revealed in south Siberia suggests that this region was a place of several expansions of hg N haplotypes at different times during the Holocene.
Materials and methods
Subjects and DNA typing
A total of 1,438 samples (whole blood and hair root samples) from unrelated males were collected in populations of south Siberia (south Altaians, Shors, Tuvinians, Tofalars, Sojots, Khakassians, Buryats, Kalmyks, Evenks, Yakuts, Evens, Koryaks), central and southwest Asia (Mongolians, Koreans, Tajiks, Iranians) and Eastern Europe (Russians) (Table 1). All samples were collected with appropriate ethical approval and informed consent.
Hg N markers LLY22g (for the whole hg N) (Jobling and Tyler-Smith 1995), Tat (for N3) (Zerjal et al. 1997), M178 (for N3a) (Underhill et al. 2000; Kharkov et al. 2005), P43 (for N2) (Karafet et al. 2002) were assayed as described in the referred papers. The Y-SNP haplogroup nomenclature used here is according to the recommendations of the Y Chromosome Consortium (2002). A total of 234 samples belonging to haplogroups N2 and N3 were analyzed at 11 STR loci (DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439) using PowerPlex® Y System (Promega Corporation, Madison, WI). Products of amplification were analyzed on ABI 3100 Genetic Analyzer (Applied Biosystems) with an appropriate filter set prepared on the basis of the relevant PowerPlex® Matrix Standards provided by Promega Corporation. Electrophoresis results were analyzed using Genscan v. 3.7 and Genotyper v. 3.7 software (Applied Biosystems). Alleles were designated by repeat numbers.
Data analysis
Median-joining (MJ) networks of hg N STR-haplotypes were constructed using the Network 4.1.1.2 program (http://www.fluxus-engineering.com). Networks were calculated by the median-joining method after processing the data with the reduced median network method in order to reduce reticulations within the network. For network construction, STR loci were weighted according to the average of their variability in the corresponding haplogroups. All of these approaches are described elsewhere (Bandelt et al. 2000).
The age of microsatellite variation within Y-chromosome haplogroups was estimated as the average squared difference in the number of repeats between all current chromosomes and the founder haplotype, averaged over microsatellite loci and divided by w = 6.9 × 10−4 per 25 years (Zhivotovsky et al. 2004). We followed the recommendation of Sengupta et al. (2006) and used the median haplotypes (formed by the median values of the repeat scores at each microsatellite locus within each haplogroup) as the founding ones.
Published hg N STR-data for Eastern Europeans (Komis, Tatars, Maris, Udmurts, Bashkirs, Karelians, Vepsas, Estonians, Ukrainians and Slovaks) and Asians (Altaians, Tuvinians, Khakassians, Evenks, Eskimo and Chinese) (Rootsi et al. 2007) were also included in the analyses.
Results and discussion
SNP analysis of 1,438 Y-chromosomes from 17 ethnic groups representing populations of north Asia and Eastern Europe demonstrates that hgs N3a and N2 are relatively frequent in some populations (Table 1). Haplogroup N3a, present at its highest frequency in Yakuts (80%), Tofalars (27%), Koryaks (25%) and Buryats (18%), is relatively frequent in all other Siberian populations as well as in the Russians (11%) studied. Haplogroup N2 chromosomes are present in the majority of the Siberian samples, reaching the highest frequencies in Tofalars (43%), Khakassians (34%) and Evenks (24%). However, this haplogroup is absent in northeastern Siberian populations (such as Evens, Koryaks and Yakuts). In addition, hg N2 is very rare in Russians (only 0.2%).
In order to obtain a better understanding of phylogenetic relationships between Y-chromosomes found in individuals carrying N2- and N3-lineages, we have determined their haplotypes at 11 microsatellite loci (Table 2). In general, 234 samples were genotyped. The age of accumulated STR variation within hg N3a estimated using the method of Zhivotovsky et al. (2004) indicates that this haplogroup is much more diverse in Eastern Europe than in Siberia (giving ages of 9.0 ky and 7.9 ky ago, respectively) (Table 3), which is in agreement with earlier observations made by Villems et al. (2002). However, median network analysis has shown that, in fact, there are two subclusters of N3a, both present in Siberia and Eastern Europe—N3a1 and N3a2 (Fig. 1). This subdivision of hg N3a has been suggested recently (Rootsi et al. 2007); however, phylogenetic and statistical analysis of hg N3a subclusters had not been performed.
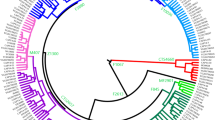
Median-joining network of haplogroup N3a based on 11 STR loci (DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439). The network includes 172 N3a-chromosomes of which 151 are novel (according to data presented in Table 2) and 21 were reported elsewhere (Rootsi et al. 2007). Each circle represents a haplotype, defined by a combination of STR markers. Circle size is shown proportional to haplotype frequency. Haplotypes are labeled as follows: Ru Russian, Yk Yakut, Tv Tuvinian, Kk Koryak, Ek Evenk, St Sojot, Mg Mongolian, Br Buryat, Al Altaian, Tf Tofalar, Kh Khakassian, Ev Even, Es Estonian, Sl Slovak, Ko Komi, Ch Chuvash, Ma Mari, Ud Udmurt, Ba Bashkir, Uk Ukrainian, Vp Vepsa, Ka Karelian. Colors indicate the subdivisions inside haplogroups: white for N3a1 haplotypes, black for N3a2 haplotypes
The presumed founder haplotypes for both subclusters (they are shown in bold in Table 2) are one-step neighbors to each other because there is only one difference in the numbers of repeats at the DYS391 locus between the two haplotypes. Nevertheless, this subdivision appears to be phylogenetically stable, because only four haplotypes (two in Russians, one in Yakuts and one in Khakassians) out of 40 N3a1-haplotypes demonstrated back-mutations from DYS392-11 to DYS392-10 allele (Table 2). Meanwhile, no back mutations from DYS392-10 to DYS392-11 allele were revealed in subcluster N3a2. Our analysis has shown that subcluster N3a1 is much older than N3a2, giving the age of 9.1 ky and 5.0 ky, respectively. Importantly, the subcluster N3a1 appears to be older in south Siberia than in Europe (10 ky and 8.2 ky ago, respectively), allowing the suggestion that the first expansion of N3a1 occurred in the south Siberian region, and then this subcluster spread to the West, into the Urals and Eastern Europe. The ancestral haplotype for N3a1 is present in Eastern Europeans (Russians and Estonians) and in south Siberians from the east Sayan regions (such as Tuvinians, Tofalars and Sojots), giving also rise to regional-specific branches of N3a1 in Yakuts and Khakassians/Tuvinians (Fig. 1). In general, this subcluster encompasses 58% of N3a individuals studied here (Table 1). It is noteworthy that the overwhelming majority of Russians (83%) as well as other Slavs and Baltic-Finnic individuals taken from the study of Rootsi et al. (2007) belong to subcluster N3a1.
Subcluster N3a2 is characterized by different tree topology. It has an almost star-like branching pattern in south Siberia, where it is present mostly in the Baikal region among Buryats. But it shows a very complex picture of the phylogenetic tree branching into its Eastern European part represented predominantly by Finno-Ugric and Turkic-speaking populations of the Volga-Ural region. High STR variation of subcluster N3a2 observed in the Volga-Ural populations, in comparison with south Siberians (6.6 ky vs. 3.7 ky ago), seems to be a consequence of serial bottlenecks and/or the small sample sizes of Eastern European N3a2 chromosomes studied. In addition, the Eastern European part of the N3a2 network displayed clear non-starlike features, which greatly reduce the precision of genetic age estimates (Sailard et al. 2000). To elucidate this question, much more detailed information on STR variation should be obtained from Finno-Ugric and Turkic-speaking populations of the Volga-Ural region. Meanwhile, traces of recent expansions of subcluster N3a2 are obvious in different populations of south Siberia—mainly, in Buryats and south Altaians.
It has been recently demonstrated that N2 haplotypes form two separate subgroups, N2-A and N2-E, characterized by an Asian and European pattern of distribution, respectively (Rootsi et al. 2007). Analyzing STR variation of N2 haplotypes in a large sample of Siberians, we have found that all of them belong to cluster N2-A. According to our data, the age of cluster N2-A is 4.8 ± 1.6 ky, which is somewhat less than estimates made by Rootsi et al. (2007). Median network analysis of N2-A haplotypes showed an existence of two subclusters determined by different STR composition in their founder haplotypes (with differences at loci DYS19, DYS391 and DYS439). Subcluster N2-A1, with founder haplotype present in different populations (such as Khakassians, Shors, Mongols, Altaians, Evenks and Tofalars), appears to be an ancestral subcluster, later giving rise to N2-A2 and N2-E. This follows from its central position in the median joining tree constructed with taking into account the location of N* lineages (Fig. 2). The age of STR variation within hg N2-A1 is a little higher than that of N2-A2 (2.4 and 1.8 kys, respectively). Interestingly, N2-A2 expansion is restricted mainly to the Tuva region.
Median-joining network of haplogroup N2 based on 11 STR loci (DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439). The network is rooted in haplogroup N*, represented by Chinese (Cn) haplotypes (Rootsi et al. 2007). The network includes 97 N2 chromosomes, of which 84 are novel (according to data presented in Table 2) and 13 were reported elsewhere (Rootsi et al. 2007). Each circle represents a haplotype, defined by a combination of STR markers. Circle size is shown proportional to haplotype frequency. Haplotypes are labeled as follows: Ru Russian, Tv Tuvinian, Ek Evenk, Mg Mongolian, Br Buryat, Al Altaian, Tf Tofalar, Kh Khakassian, Sh Shor, Km Kalmyk, Em Eskimo, Ta Tatar, Ko Komi, Ma Mari, and Vp Vepsa
Overall, the data obtained indicate that south Siberia was a place of several expansions of hgs N2 and N3 haplotypes at different times during the Holocene. Future examinations of Y-chromosome variation using a combined STR-SNP approach in additional population samples may enable a better definition of the differences between north Eurasian population groups.
References
Bandelt HJ, Macaulay V, Richards M (2000) Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol Phylogenet Evol 16:8–28
Cinnioglu C, King R, Kivisild T, Kalfoglu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA (2004) Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 114:127–148
Jobling MA, Tyler-Smith C (1995) Fathers and sons: the Y chromosome and human evolution. Trends Genet 111:449–456
Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF (2002) High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol 74:761–789
Kharkov VN, Stepanov VA, Feshchenko SP, Borinskaya SA, Yankovsky NK, Puzyrev VP (2005) Frequencies of Y chromosome binary haplogroups in Belarussians. Russ J Genet 41:928–931
Rootsi S, Zhivotovsky LA, Baldovic M, Kayser M, Kutuev IA, Khusainova R, Bermisheva MA, Gubina M, Fedorova SA, Ilumae AM, Khusnutdinova EK, Voevoda MI, Osipova LP, Stoneking M, Lin AA, Ferak V, Parik J, Kivisild T, Underhill PA, Villems R (2007) A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet 15:204–211
Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Côrte-Real HBSM, Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, Gölge M, Hill E, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenco SA, Krumina A, Kučinskas V, Lavinha J, Livshits LA, Malaspina P, Maria S, McElreavey K, Meitinger TA, Mikelsaar AV, Mitchell RJ, Nafa K, Nicholson J, Nørby S, Pandya A, Parik J, Patsalis PC, Pereira L, Peterlin B, Pielberg G, Prata MJ, Previderé C, Roewer L, Rootsi S, Rubinsztein DC, Saillard J, Santos FR, Stefanescu G, Sykes BC, Tolun A, Villems R, Tyler-Smith C, Jobling MA (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543
Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726
Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE, Lin AA, Mitra M, Sil SK, Ramesh A, Usha Rani MV, Thakur CM, Cavalli-Sforza LL, Majumder PP, Underhill PA (2006) Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of central Asian pastoralists. Am J Hum Genet 78:202–221
Underhill PA (2003) Inferring human history: clues from Y-chromosome haplotypes. Cold Spring Harb Symp Quant Biol 68:487–493
Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361
Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62
Villems R, Rootsi S, Tambets K, Adojaan M, Orekhov V, Khusnutdinova E, Yankovsky N (2002) Archaeogenetics of Finno-Ugric-speaking populations. In: Julku K (ed) The roots of peoples and languages of the Northern Eurasia IV. Societas Historiae Fenno-Ugricae, Oulu pp 271–284
Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2:339–348
Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183
Zhivotovsky LA, Underhill PA, Cinnioglu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L (2004) The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 74:50–61
Acknowledgments
We are grateful to all the voluntary donors of DNA samples used in this study and to C. Dorzhu for Tuvinian samples, F. Luzina for Altaian and Shor samples, H. Lee for Korean samples, T. Tsedev for Mongolian samples, and J. Rostamzadeh for Persian and Kurd samples. We thank A. Jakubowska and M. Perkova for technical assistance. This study was supported by the Russian Foundation for Basic Research grants 06-04-48136 (to B.M.), 07-04-00445 (to M.D.), Polish State Committee for Scientific Research grant 3P04C 04823 (to T.G.), Far-East Branch of the Russian Academy of Sciences grant 06-3-B-06-216 (to G.D.), and Program of Basic Research of Russian Academy of Sciences “Biodiversity and Dynamics of Gene Pools” (to B.M.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Derenko, M., Malyarchuk, B., Denisova, G. et al. Y-chromosome haplogroup N dispersals from south Siberia to Europe. J Hum Genet 52, 763–770 (2007). https://doi.org/10.1007/s10038-007-0179-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-007-0179-5
Keywords
This article is cited by
-
Genetic insights into the social organisation of the Avar period elite in the 7th century AD Carpathian Basin
Scientific Reports (2020)
-
Mitogenomic diversity and differentiation of the Buryats
Journal of Human Genetics (2018)
-
A new self-learning computational method for footprints of early human migration processes
Molecular Genetics and Genomics (2018)
-
Y chromosome haplotype diversity in Mongolic-speaking populations and gene conversion at the duplicated STR DYS385a,b in haplogroup C3-M407
Journal of Human Genetics (2016)
-
Y-SNP L1034: limited genetic link between Mansi and Hungarian-speaking populations
Molecular Genetics and Genomics (2015)