Abstract
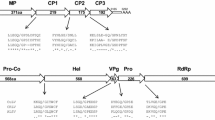
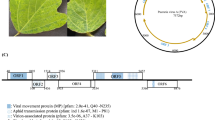
The complete genome sequences of RNA1 and RNA2 of the oca strain of the potato virus arracacha virus B were determined using next-generation sequencing. The RNA1 molecule is predicted to encode a 259-kDa polyprotein with homology to proteins of the cheraviruses apple latent spherical virus (ALSV) and cherry rasp leaf virus (CRLV). The RNA2 molecule is predicted to encode a 102-kDa polyprotein which also has homology to the corresponding protein of ALSV and, to a lesser degree, CRLV (30 % for RNA1, 24 % for RNA2). Detailed analysis of the genome sequence confirms that AVB is a distinct member of the genus Cheravirus.


Similar content being viewed by others
References
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2010) GenBank. Nucleic Acids Res 38:D46–D51
CABI, EPPO Arracacha B ‘nepovirus’, oca strain, Data Sheets on Quarantine Pests, Prepared by CABI and EPPO for the EU under Contract 90/399003. http://www.eppo.org/QUARANTINE/virus/Arracacha_B_virus/AVBO00_ds.pdf
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421
Candresse T, Svanella-Dumas L, Le Gall O (2006) Characterization and partial genome sequence of stocky prune virus, a new member of the genus Cheravirus. Arch Virol 151:1179–1188
EPPO (1984) Fitches informatives OEPP sur les organismes de quarataine; Potato Viruses (non-European). EPPO Bull 14:11–22
Foster GD, Taylor S (1998) Plant virology protocols: from virus isolation to transgenic resistance. Humana Press, New York
Gorbalenya AE, Donchenko AP, Blinov VM, Koonin EV (1989) Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett 243:103–114
Gorbalenya AE, Koonin EV, Wolf YI (1990) A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett 262:145–148
Hellen CUT, Kraeusslich HG, Wimmer E (1989) Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry 28:9881–9890
James D, Howell WE, Mink GI (2001) Molecular evidence of the relationship between a virus associated with flat apple Disease and cherry rasp leaf virus as determined by RT-PCR. Plant Dis 85:47–52
James D, Upton C (2002) Nucleotide sequence analysis of RNA-2 of a flat apple isolate of Cherry rasp leaf virus with regions showing greater identity to animal picornaviruses than to related plant viruses. Arch Virol 147:1631–1641
Jones RAC (1981) OCA strain of arracacha virus-B from potato in Peru. Plant Dis 65:753–754
Jones RAC, Kenten RH (1981) A strain of arracacha virus-B infecting Oca (Oxalis Tuberosa, Oxalidaceae) in the Peruvian Andes. Phytopathologische Zeitschrift: J Phytopathol 100:88–95
Kenten RH, Jones RAC (1979) Arracacha virus B, a second isometric virus infecting arracacha (Arracacia xanthorrhiza; Umbelliferae) in the Peruvian Andes. Ann Appl Biol 93:31–36
Koonin EV (1991) The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72(Pt 9):2197–2206
Koonin EV, Mushegian AR, Ryabov EV, Dolja VV (1991) Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J Gen Virol 72(Pt 12):2895–2903
Le Gall O, Sanfaçon H, Ikegami M, Iwanami T, Jones T, Karasev A, Lehto K, Wellink J, Wetzel T, Yoshikawa N (2007) Cheravirus and Sadwavirus: two unassigned genera of plant positive-sense single-stranded RNA viruses formerly considered atypical members of the genus Nepovirus (family Comoviridae). Arch Virol 152:1767–1774
Li C, Yoshikawa N, Takahashi T, Ito T, Yoshida K, Koganezawa H (2000) Nucleotide sequence and genome organization of Apple latent spherical virus: a new virus classified into the family Comoviridae. J Gen Virol 81:541–547
Maliogka V, Dovas CI, Efthimiou K, Katis NI (2004) Detection and differentiation of Comoviridae species using a semi-nested RT-PCR and a phylogenetic analysis based on the polymerase protein. J Phytopathol 152:404–409
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D (2010) Tablet-next generation sequence assembly visualization. Bioinformatics 26:401–402
Ritzenthaler C, Viry M, Pinck M, Margis R, Fuchs M, Pinck L (1991) Complete nucleotide sequence and genetic organization of grapevine fanleaf nepovirus RNA1. J Gen Virol 72(Pt 10):2357–2365
Sanfaçon H, Wellink J, Le Gall O, Karasev A, van der Vlugt R, Wetzel T (2009) Secoviridae: a proposed family of plant viruses within the order Picornavirales; that combines the families Sequiviridae; and Comoviridae, the unassigned genera Cheravirus and Sadwavirus, and the proposed genus Torradovirus. Arch Virol 154:899–907
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JR, Perry KL, De Jong W (2004) A new potato virus in a new lineage of picorna-like viruses. Arch Virol 149:2141–2154
Acknowledgment
This work was funded by the EU Qbol project and Defra
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Adams, I.P., Glover, R., Souza-Richards, R. et al. Complete genome sequence of arracacha virus B: a novel cheravirus. Arch Virol 158, 909–913 (2013). https://doi.org/10.1007/s00705-012-1546-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-012-1546-x