Abstract
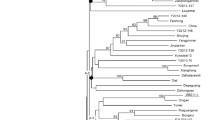
Bananas in Thailand have been surveyed by our team to be at least 140 cultivars in the plantations, 10 wild species and, 4 introduced species. To characterize the genetic relationship of species and cultivars, a set of novel SSR markers was developed. Totaling 53 clones containing SSR motifs were isolated from SSR-enriched library of wild Musa balbisiana Colla ‘Tani’ (BB). Selected positive clones were used to design 28 primer pairs for amplification of 12 wild and 82 cultivar accessions with genome designations AA, AB, AAA, AAB, ABB, and BBB. These SSR markers loci were homology searched to the banana genomes to map their locations. The seven-sets multiplex PCR approach using four fluorescent-labeled universal primers were utilized for cost effectiveness. Capillary fragment analysis yielded the accurate size of amplicons for evaluation of particular patterns for each cultivar. Phylogram and Structure analysis presented the specific genotype of genome groups (A and B genotypes, polyploid hybrid genomes) and cultivar groups. By A:B specific alleles ratio, accurate genome designations of hybrids can be determined. Additionally, a marker, characterized to be partial plastid ycf2 gene, indicated the maternal identification of hybrid cultivars. One SSR marker was also preliminary tested with some wild species and advised to be the candidate fingerprinting marker for species identification. In conclusion, SSR marker sets developed here proved their exploitation in detailed identity and relationship of cultivated bananas, which would be useful for genetic conservation and ongoing breeding programs in Thailand and other areas.





Similar content being viewed by others
References
Addisalem AB, Esselink GD, Bongers F, Smulders MJM (2015) Genomic sequencing and microsatellite marker development for Boswellia papyrifera, an economically important but threatened tree native to dry tropical forests. AoB PLANTS 7:plu086. doi:10.1093/aobpla/plu086
Anderson JA, Churchill GA, Sutrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186. doi:10.1139/g93-024
Barrett CF, Specht CD, Leebens-Mack J, Stevenson DW, Zomlefer WB, Davis JI (2014) Resolving ancient radiation: can complete plastid gene sets elucidate deep relationships among the tropical gingers (Zingiberales)? Ann Bot (Oxford) 113:119–133. doi:10.1093/aob/mct264
Bartoš J, Alkhimova O, Doleželová M, De Langhe E, Doležel J (2005) Nuclear genome size and genomic distribution of ribosomal DNA in Musa and Ensete (Musaceae): taxonomic implications. Cytogenet Genome Res 109:50–57. doi:10.1159/000082381
Biémont C (2008) Genome size evolution: within-species variation in genome size. Heredity 101:297–298. doi:10.1038/hdy.2008.80
Blacket MJ, Robin C, Good RT, Lee SF, Miller AD (2012) Universal primers for fluorescent labelling of PCR fragments—an efficient and cost-effective approach to genotyping by fluorescence. Molec Ecol Resources 12:456–463. doi:10.1111/j.1755-0998.2011.03104.x
Bowers JE, Bachlava E, Brunick RL, Rieseberg LH, Knapp SJ, Burke JM (2012) Development of a 10,000 locus genetic map of the sunflower genome based on multiple crosses. G3 (Bethesda) 2:721–729. doi:10.1534/g3.112.002659
Buhariwalla HK, Jarret RL, Jayashree B, Crouch JH, Oritiz R (2005) Isolation and characterization of microsatellite markers from Musa balbisiana. Molec Ecol Notes 5:327–330. doi:10.1111/j.1471-8286.2005.00916.x
Chomchalow N, Silayoi B (1984) Banana germplasm in Thailand. IBPGR/SEAP Newslett 8:23–28. http://pikul.lib.ku.ac.th/Fulltext_kukr/KU0222046c.pdf
Christelová P, Valárik M, Hřibová E, Van den Houwe I, Channelière S, Roux N, Doležel J (2011) A platform for efficient genotyping in Musa using microsatellite markers. AoB Plants 2011:plr024. doi:10.1093/aobpla/plr024
Clark LV, Jasieniuk M (2011) POLYSAT: an R package for polyploid microsatellite analysis. Molec Ecol Resources 11:562–566. doi:10.1111/j.1755-0998.2011.02985.x
Creste S, Benatti TR, Orsi MR, Risterucci A-M, Figueira A (2006) Isolation and characterization of microsatellite loci from a commercial cultivar of Musa acuminata. Molec Ecol Notes 6:303–306. doi:10.1111/j.1471-8286.2005.01209.x
Crouch JH, Crouch HK, Ortiz R, Jarret RL (1997) Microsatellite markers for molecular breeding of Musa. InfoMusa 6:5–6. doi:10.1300/J144v01n01_06
Culley TM, Stamper TI, Stokes RL, Brzyski JR, Hardiman NA, Klooster MR, Merritt BJ (2013) An efficient technique for primer development and application that integrates fluorescent labeling and multiplex PCR. Appl Plant Sci 1:1300027. doi:10.3732/apps.1300027
Davey MW, Gudimella R, Harikrishna JA, Sin LW, Khalid N, Keulemans J (2013) A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genom 14:683. doi:10.1186/1471-2164-14-683
de Jesus ON, Silva SDOE, Amorim EP, Ferreira CF, de Campos JMS, Silva GDG, Figueira A (2013) Genetic diversity and population structure of Musa accessions in ex situ conservation. BMC Plant Biol 13:41. doi:10.1186/1471-2229-13-41
Delourme R, Falentin C, Fomeju BF et al (2013) High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genom 14:120. doi:10.1186/1471-2164-14-120
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079. doi:10.1101/gr.132102
D’Hont A, Paget-Goy A, Escoute J, Carreel F (2000) The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theor Appl Genet 100:177–183. doi:10.1007/s001220050024
D’Hont A, Denoeud F, Aury J-M et al (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217. doi:10.1038/nature11241
Doležel J, Doleželová M, Novák FJ (1994) Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol Plant 36:351–357
Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protocols 2:2233–2244. doi:10.1038/nprot.2007.310
Droc G, Larivière D, Guignon V et al (2013) The banana genome hub. Database (Oxford) 2013:035. doi:10.1093/database/bat035
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resources 4:359–361. doi:10.1007/s12686-011-9548-7
Esselink GD, Nybom H, Vosman B (2004) Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting—peak ratios) method. Theor Appl Genet 109:402–408. doi:10.1007/s00122-004-1645-5
Eustice M, Yu Q, Lai CW, Hou S, Thimmapuram J, Liu L, Alam M, Moore PH, Presting GG, Ming R (2008) Development and application of microsatellite markers for genomic analysis of papaya. Tree Genet Genomes 4:333–341. doi:10.1007/s11295-007-0112-2
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molec Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molec Ecol Notes 7:574–578. doi:10.1111/j.1471-8286.2007.01758.x
Fredotović Z, Šamanić I, Weiss-Schneeweiss H, Kamenjarin J, Jang T, Puizina J (2014) Triparental origin of triploid onion, Allium × cornutum (Clementi ex Visiani, 1842), as evidenced by molecular, phylogenetic and cytogenetic analyses. BMC Plant Biol 14:24. doi:10.1186/1471-2229-14-24
Ganal MW, Durstewitz G, Polley A et al (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLOS One 6:e28334. doi:10.1371/journal.pone.0028334
Ge XJ, Liu MH, Wang WK, Schaal BA, Chiang TY (2005) Population structure of wild bananas, Musa balbisiana, in China determined by SSR fingerprinting and cpDNA PCR-RFLP. Molec Ecol 14:933–944. doi:10.1111/j.1365-294x.2005.02467.x
Ge C, Cui YN, Jing PY, Hong XY (2014) An alternative suite of universal primers for genotyping in multiplex PCR. PLOS One 9:e92826. doi:10.1371/journal.pone.0092826
Getachew S, Mekbib F, Admassu B, Kelemu S, Kidane S, Negisho K, Djikeng A, Nzuki I (2014) A look into genetic diversity of Enset (Ensete ventricosum (Welw.) Cheesman) using transferable microsatellite sequences of banana in Ethiopia. J Crop Improv 28:159–183. doi:10.1080/15427528.2013.861889
Häkkinen M (2013) Reappraisal of sectional taxonomy in Musa (Musaceae). Taxon 62:809–813. doi:10.12705/624.3
Häkkinen M, Hong W (2007) New species and variety of Musa (Musaceae) from Yunnan, China. Novon 17:440–446. doi:10.3417/1055-3177(2007)17[440:NSAVOM]2.0.CO;2
Heslop-Harrison JS, Schwarzacher T (2007) Domestication, genomics and the future for banana. Ann Bot (Oxford) 100:1073–1084. doi:10.1093/aob/mcm191
Hippolyte I, Bakry F, Seguin M et al (2010) A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biol 10:65. doi:10.1186/1471-2229-10-65
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877. doi:10.1101/gr.9.9.868
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800. doi:10.1038/nature03895
Iwata H, Kato T, Ohno S (2000) Triparental origin of Damask roses. Gene 259:53–59. doi:10.1016/S0378-1119(00)00487-x
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi:10.1093/bioinformatics/btm233
James EA, Brown GK, Citroen R, Rossetto M, Porter C (2011) Development of microsatellite loci in Triglochin procera (Juncaginaceae), a polyploid wetland plant. Conservation Genet Resources 3:103–105. doi:10.1007/s12686-010-9301-7
Jeridi M, Bakry F, Escoute J, Fondi E, Carreel F, Ferchichi A, D’Hont A, Rodier-Goud M (2011) Homoeologous chromosome pairing between the A and B genomes of Musa spp. revealed by genomic in situ hybridization. Ann Bot (Oxford) 108:975–981. doi:10.1093/aob/mcr207
Kaemmer D, Fischer D, Jarret RL, Baurens F-C, Grapin A, Dambier D, Noyer J-L, Lanaud C, Kahl G, Lagoda PJL (1997) Molecular breeding in the genus Musa: a strong case for STMS marker technology. Euphytica 96:49–63. doi:10.1023/A:1002922016294
Lagoda PJ, Noyer JL, Dambier D, Baurens FC, Grapin A, Lanaud C (1998) Sequence tagged microsatellite site (STMS) markers in the Musaceae. Molec Ecol 7:659–663
Lysák MA, Doleželová M, Horry JP, Swennen R, Doležel J (1999) Flow cytometric analysis of nuclear DNA content in Musa. Theor Appl Genet 98:1344–1350
Martin G, Baurens FC, Cardi C, Aury J, D’Hont A (2013) The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PLOS One 8:e67350. doi:10.1371/journal.pone.0067350
Mbanjo EGN, Tchoumbougnang F, Mouelle AS, Oben JE, Nyine M, Dochez C, Ferguson ME, Lorenzen J (2012) Molecular marker-based genetic linkage map of a diploid banana population (Musa acuminata Colla). Euphytica 188:369–386. doi:10.1007/s10681-012-0693-1
Miller RNG, Passos MAN, Menezes NNP, Souza MT, Costa MMC, Azevedo VCR, Amorim EP, Pappas GJ, Ciampi AY (2010) Characterization of novel microsatellite markers in Musa acuminata subsp. burmannicoides, var. Calcutta 4. BMC Res Notes 3:148. doi:10.1186/1756-0500-3-148
Missiaggia A, Grattapaglia D (2006) Plant microsatellite genotyping with 4-color fluorescent detection using multiple-tailed primers. Genet Molec Res 5:72–78
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nsabimana A, van Staden J (2006) Ploidy investigation of bananas (Musa spp.) from the National Banana Germplasm Collection at Rubona-Rwanda by flow cytometry. S African J Bot 72:302–305. doi:10.1016/j.sajb.2005.10.004
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. CIRAD Agricultural Research for Development. http://darwin.cirad.fr/
Pillay M, Ogundiwin E, Nwakanma DC, Ude G, Tenkouano A (2001) Analysis of genetic diversity and relationships in East African banana germplasm. Theor Appl Genet 102:965–970. doi:10.1007/s001220000500
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rahman AY, Usharraj AO, Misra BB et al (2013) Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genom 14:75. doi:10.1186/1471-2164-14-75
Ravishankar KV, Raghavendra KP, Athani V, Rekha A, Sudeepa K, Bhavya D, Srinivar V, Ananad L (2013) Development and characterisation of microsatellite markers for wild banana (Musa balbisiana). J Hort Sci Biotechnol 88:605–609
Ray T, Dutta I, Saha P, Das S, Roy SC (2006) Genetic stability of three economically important micropropagated banana (Musa spp.) cultivars of lower Indo-Gangetic plains, as assessed by RAPD and ISSR markers. Plant Cell Tissue Organ Cult 85:11–21. doi:10.1007/s11240-005-9044-4
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molec Biol Evol 4:406–425
Schlötterer C, Harr B (2004) Microsatellite instability. Wiley, Hoboken, NJ, USA. doi:10.1038/npg.els.0000840
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115. doi:10.1126/science.1178534
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234. doi:10.1038/72708
Shokeen B, Sethy NK, Kumar S, Bhatia S (2007) Isolation and characterization of microsatellite markers for analysis of molecular variation in the medicinal plant Madagascar periwinkle (Catharanthus roseus (L.) G. Don). Plant Sci (Elsevier) 172:441–451. doi:10.1016/j.plantsci.2006.10.010
Simmonds NW (1962) The evolution of the bananas. Longmans, London
Simmonds NW, Shepherd K (1955) The taxonomy and origins of the cultivated bananas. J Linn Soc Bot 55:302–312. doi:10.1111/j.1095-8339.1955.tb00015.x
Soto JC, Ortiz JF, Perlaza-Jiménez L, Vásquez AX, Lopez-Lavalle LA, Mathew B, Léon J, Bernal AJ, Ballvora A, López CE (2015) A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes. BMC Genom 16:190. doi:10.1186/s12864-015-1397-4
Swangpol S, Somana J (2011) Musa serpentina (Musaceae): a new banana species from western border of Thailand. Thai Forest Bull Bot 39:31–36
Swangpol S, Volkaert HA, Sotto RC, Seelanan T (2007) Utility of selected non-coding chloroplast DNA sequences for lineage assessment of Musa interspecific hybrids. J Biochem Molec Biol 40:577–587
Swangpol S, Somana J, Chattrakom S, Sukkaewmanee N, Kiatprapai P (2009) 108 Thai Banana Cultivars [in Thai]. Bangkok Publishing, Bangkok
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452. doi:10.1101/gr.184001
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. doi:10.1038/35048692
The Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195. doi:10.1038/nature10158
Thomson MJ, Zhao K, Wright M et al (2012) High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Molec Breed 29:875–886. doi:10.1007/s11032-011-9663-x
Valmayor RV, Jamaluddin SH, Silayoi B, Kusumo S, Danh LD, Pascua OC, Espino RRC (2000) Banana cultivar names and synonyms in Southeast Asia. International Network for Improvement of Banana and Plantain—Asia and the Pacific Office, Los Baños
van Dijk T, Noordijk Y, Dubos T, Bink MC, Meulenbroek BJ, Visser RG, van de Weg E (2012) Microsatellite allele dose and configuration establishment (MADCE): an integrated approach for genetic studies in allopolyploids. BMC Plant Biol 12:25. doi:10.1186/1471-2229-12-25
Vukosavljev M, Esselink GD, Van′t Westende WPC, Cox P, Visser RGF, Arens P, Smulders MJM (2015) Efficient development of highly polymorphic microsatellite markers based on polymorphic repeats in transcriptome sequences of multiple individuals. Molec Ecol Resources 15:17–27. doi:10.1111/1755-0998.12289
Wang JY, Zheng LS, Huang BZ, Liu WL, Wu YT (2010) Development, characterization, and variability analysis of microsatellites from a commercial cultivar of Musa acuminata. Genet Resources Crop Evol 57:553–563. doi:10.1007/s10722-009-9493-4
Weising K, Nybom H, Wolff K, Kahl G (2005) DNA fingerprinting in plants: principles, methods, and applications, 2nd edn. CRC Press, Boca Raton
Wongniam S, Somana J, Swangpol S, Seelanan T, Chareonsap P, Chadchawan S, Jenjittikul T (2010) Genetic diversity and species-specific PCR-based markers from AFLP analyses of Thai bananas. Biochem Syst Ecol 38:416–427. doi:10.1016/j.bse.2010.03.015
Zalapa JE, Cuevas H, Zhu H, Steffan S, Senalik D, Zeldin E, McCown B, Harbut R, Simon P (2012) Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Amer J Bot 99:193–208. doi:10.3732/ajb.1100394
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Molec Ecol 11:1–16. doi:10.1046/j.0962-1083.2001.01418.x
Acknowledgments
This research was financially supported by the Thailand Research Fund in collaboration with the Commission on Higher Education of Thailand to JS (MRG5380133) and to SCS (MRG5280100) and Plant Genetic Conservation Project, under the Royal Initiative of Her Royal Highness Princess Maha Chakri Sirindhorn to PPC. Research Assistant Supporting Grant was provided by Faculty of Science, Mahidol University to TR. The authors are grateful for banana samples and knowledge from all institutes and collectors including Pra Sobhon Khanaporn and Mr. Samadchai Chattrakhom.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Eric Schranz.
Below is the link to the electronic supplementary material.
606_2015_1274_MOESM1_ESM.doc
Online Resource 1 Banana samples, 12 wild and 82 cultivar accessions from the collections in Thailand, with their source locations (DOC 166 kb)
606_2015_1274_MOESM4_ESM.doc
Online Resource 4 SSR loci information and position on designated chromosomes of AA genome: Musa acuminata accession ‘Pahang’ (DOC 246 kb)
606_2015_1274_MOESM5_ESM.doc
Online Resource 5 SSR loci information and position on designated chromosomes of BB genome: Musa balbisiana accession ‘Pisung Klutuk Wulung’ (DOC 237 kb)
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Banana samples, 12 wild and 82 cultivar accessions from the collections in Thailand, with their source locations.
Online Resource 2. Supplementary materials and methods.
Online Resource 3. List of banana cultivars in Thailand with previous and suggested genome designations.
Online Resource 4. SSR loci information and position on designated chromosomes of AA genome: Musa acuminata accession ‘Pahang’.
Online Resource 5. SSR loci information and position on designated chromosomes of BB genome: Musa balbisiana accession ‘Pisung Klutuk Wulung’.
Online Resource 6. Microsatellite profiling of A- and B-genome specific alleles in 17 SSR markers.
Online Resource 7. Estimation of optimal K evaluated by Structure Harvester program.
Rights and permissions
About this article
Cite this article
Rotchanapreeda, T., Wongniam, S., Swangpol, S.C. et al. Development of SSR markers from Musa balbisiana for genetic diversity analysis among Thai bananas. Plant Syst Evol 302, 739–761 (2016). https://doi.org/10.1007/s00606-015-1274-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-015-1274-2