Abstract
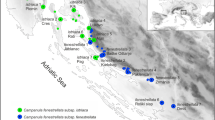
The high-altitude Southeastern Brazilian Cattleya coccinea clade includes two taxonomically challenging species, Cattleya coccinea and Cattleya mantiqueirae, the latter considered restricted to the mountain range of Serra da Mantiqueira. To hypothesize the existence of evolutionary independent lineages corresponding to these species, we inferred phylogenetic relationships and described patterns of population genetic diversity and structure of populations from six localities based on ISSR and cpDNA sequence data. Results do not support the species C. coccinea and C. mantiqueirae as previously circumscribed. Most analyses recovered southwestern and northeastern groups along the two mountain ranges, suggesting geography rather than morphology as an important criteria for species delimitation. However, due to the lack of information on the type locality of C. coccinea and the topological complexity of the southwestern area, the taxonomic circumscription of C. coccinea and C. mantiqueirae must be evaluated by additional population sampling. We also propose that the northeastern group comprise at least one distinct species. Also, specimens from Lima Duarte from Serra da Mantiqueira clearly do not belong to neither of the two groups and demand further investigation concerning a possible hybridization scenario of C. coccinea/C. mantiqueirae and C. brevipedunculata. Population genetic patterns of variation are in agreement with pollination observations for this group and with a drift-selection model of speciation. Owing to the high genetic differences among populations, with low levels of variation within populations, conservation priorities should favour protecting as many populations as possible.





Similar content being viewed by others
References
Akaike H (1974) A new look at the statistical model identification. IEEE T Automat Contr 19:716–723
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Molec Phylogen Evol 29:417–434. doi:10.1016/S1055-7903(03)00208-2
Backeljau T, de Bruyn L, de Wolf H, Jordaens K, Van Dongan S, Verhagen R, Winnepenninckx B (1995) Random amplified polymorphic DNA (RAPD) and parsimony methods. Cladistics 11:119–130
Bailey CD, Carr TG, Harris SA, Hughes CE (2003) Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molec Phylogen Evol 29:435–455. doi:10.1016/j.ympev.2003.08.021
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Molec Phylogen Evol 1:3–16
Bateman RM, James KE, Luo YB, Lauri RK, Fulcher T, Cribb PJ, Chase MW (2009) Molecular phylogenetics and morphological reappraisal of the Platanthera clade (Orchidaceae: Orchidinae) prompts expansion of the generic limits of Galearis and Platanthera. Ann Bot (Oxford) 104:431–445. doi:10.1093/aob/mcp089
Borba EL, Funch RR, Ribeiro PL, Smidt EC, Silva-Pereira V (2007a) Demography, genetic and morphological variability of the endangered Sophronitis sincorana (Orchidaceae) in the Chapada Diamantina, Brazil. Pl Syst Evol 267:129–146. doi:10.1007/s00606-007-0555-9
Borba EL, Funch RR, Ribeiro PL, Smidt EC, Silva-Pereira V (2007b) Demografia, variabilidade genética e morfológica e conservação de Cattleya tenuis (Orchidaceae), espécie ameaçada de extinção da Chapada Diamantina. Sitientibus Sér Ci Biol 7:211–222
Braga PIS (1977) Aspectos biológicos das Orchidaceae de uma campina de Amazônia Central. Acta Amazonica 2:1–89
Bussell JD, Waycott M, Chappill JA (2005) Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspect Pl Ecol Evol Syst 7:3–26. doi:10.1016/j.ppees.2004.07.001
Chiron GR, Oliveira RP, Santos TM, Bellvert F, Bertrand C, van den Berg C (2009) Phylogeny and evolution of Baptistonia (Orchidaceae, Oncidiinae) based on molecular analyses, morphology and floral oil evidences. Pl Syst Evol 281:35–49. doi:10.1007/s00606-009-0181-9
Cogniaux CA (1901) Orchidaceae. In: Martius CFP, Eichler AG, Urban I (eds) Flora Brasiliensis 3 part 5. Monachii typographia regia, München, pp 313–324
Cozzolino S, Widmer A (2005) Orchid diversity: an evolutionary consequence of deception? Trends Ecol Evol 20:487–494. doi:10.1016/j.tree.2005.06.004
Cruz DT, Selbach-Schnadelbach A, Lambert SM, Ribeiro PL, Borba EL (2011) Genetic and morphological variability in Cattleya elongata Barb. Rodr. (Orchidaceae), endemic to the campo rupestre vegetation in northeastern Brazil. Pl Syst Evol 294:87–98. doi:10.1007/s00606-011-0444-0
de Queiroz K (1998) The general lineage concept of species, species criteria, and the process of speciation: A conceptual unification and terminological recommendations. In: Howard DJ, Berlocher SH (eds) Endless forms: Species and speciation. Oxford University Press, New York, pp 57–75
de Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886. doi:10.1080/10635150701701083
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molec Ecol 4:129–134
Descourtilz JT (1768) Cattleya coccinea Lindl. In: Epidendres des forêts vierges du Brésil. Tome 1, Bibliothèque de l’Institut de France, Paris
Devey DS, Bateman RM, Fay MF, Hawkins JA (2008) Friends or relatives? Phylogenetics and species delimitation in the controversial European orchid genus Ophrys. Ann Bot London (Oxford) 101:385–402. doi:10.1093/aob/mcm299
Dodson CH (1962) The importance of pollination in the evolution of the orchids of tropical America. Amer Orchid Soc Bull 31:525–534
Dodson CH (1965) Agentes de polinización y su influencia sobre la evolución en la familia Orquidaceae. UNAP, Iquitos, pp 45–50
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytoch Bull 19:11–15
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molec Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from matrix distances among DNA haplotypes: application to human mitochondria DNA restriction data. Genetics 131:479–491
Excoffier L, Laval LG, Schneider S (2005) ARLEQUIN, version 3.0: an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Fajardo CG, Vieira FA, Molina WF (2014) Interspecific genetic analysis of orchids in Brazil using molecular markers. Pl Syst Evol. doi:10.1007/s00606-014-1009-9
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molec Ecol Notes 7:574–578. doi:10.1111/j.1471-8286.2007.01758.x
Farris JS (1977) Phylogenetic analysis under Dollo’s law. Syst Zool 26:77–88
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Forrest AD, Hollingsworth ML, Hollingsworth PM, Sydes C, Bateman R (2004) Population genetic structure in European populations of Spiranthes romanzoffiana set in the context of other genetic studies on orchids. Heredity 92:218–227. doi:10.1038/sj.hdy.6800399
Forster W, Souza VC (2013) Laeliinae (Orchidaceae) do parque nacional do Caparaó, Estados do Espírito Santo e Minas Gerais, Brasil. Hoehnea 40:701–726
Fowlie JA (1968) Sophronitis coccinea Lindl. Orchid Digest 32:272–273
Fowlie JA (1972) A contribution to a further clarification of the genus Sophronitis Lindl., including the elevation to full specific status two previously described varieties. Orchid Digest 36:181–194
Fowlie JA (1975) With Ghillany in Brazil. Part IX. A new Sophronitis from Pico do Açu, Sophronitis acuensis. Orchid Digest 39:147–151
Fowlie JA (1987) A contribution to a monographic revision of the genus Sophronitis Lindl. Orchid Digest 51:15–32
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Gentry AH, Dodson CH (1987) Diversity and biogeography of neotropical vascular epiphytes. Ann Missouri Bot Gard 74:205–233
George S, Sharma J, Yadon VL (2009) Genetic diversity of the endangered and narrow endemic Piperia yadonii (Orchidaceae) assessed with ISSR polymorphisms. Amer J Bot 96:2022–2030. doi:10.3732/ajb.0800368
Givnish TJ, Bean GJ, Ames M, Lyon SP, Sytsma KJ (2013) Phylogeny, floral evolution, and inter-island dispersal in Hawaiian Clermontia (Campanulaceae) based on ISSR variation and plastid spacer sequences. PLoS One 8:e62566. doi:10.1371/journal.pone.0062566
Goloboff PA (1999) Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics 15:415–428
Goloboff PA, Farris JS, Nixon KC (2003) TNT: tree analysis using new technology. Program and documentation. http://www.zmuc.dk/public/phylogeny/TNT. Accessed 26 July 2012
Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24:774–786. doi:10.1111/j.1096-0031.2008.00217.x
Hammer Ø, Harper DAT, Ryan PD (2001) Past: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Hamrick JL, Godt MJW (1996) Effect of life history traits on genetic diversity in plant species. Phil Trans R Soc B 351:1291–1298
Hart MW (2011) The species concept as an emergent property of population biology. Evolution 65:613–616. doi:10.1111/j.1558-5646.2010.01202.x
Hausdorf B, Hennig C (2010) Species delimitation using dominant and codominant multilocus markers. Syst Biol 59:491–503. doi:10.1093/sysbio/syq039
Hoehne FC (1949) Iconografia de Orchidaceas do Brasil. Secretaria de Agricultura, São Paulo
Hopkins MJG (2007) Modelling the known and unknown plant biodiversity of the Amazon basin. J Biogeogr 34:1400–1411. doi:10.1111/j.1365-2699.2007.01737.x
Huelsenbeck JP, Andolfatto P (2007) Inference of population structure under a Dirichlet process model. Genetics 175:1787–1802. doi:10.1534/genetics.106.061317
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754–755. doi:10.1093/bioinformatics/btg180
Huelsenbeck JP, Andolfatto P, Huelsenbeck ET (2011) Structurama: Bayesian inference of population structure. Evol Bioinform Online 7:55–59. doi:10.4137/EBO.S6761
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi:10.1093/bioinformatics/btm233
Janes JK, Steane DA, Vaillancourt RE (2012) What does population structure analysis reveal about the Pterostylis longifolia complex (Orchidaceae)? Ecol Evol 2:2631–2644. doi:10.1002/ece3.376
Jersáková J, Johnson SD, Kindlmann P (2006) Mechanisms and evolution of deceptive pollination in orchids. Biol Rev 81:219–235. doi:10.1017/S1464793105006986
Johnson SD, Nilsson LA (1999) Pollen carryover, geitonogamy, and the evolution of deceptive pollination systems in orchids. Ecology 80:2607–2619
Kay KM, Reeves PA, Olmstead RG, Schemske DW (2005) Rapid speciation and the evolution of hummingbird pollination in neotropical Costus subgenus Costus (Costaceae): evidence from nrDNA ITS and ETS sequences. Amer J Bot 92:1899–1910. doi:10.3732/ajb.92.11.1899
Kramer AT, Fant JB, Ashley MV (2011) Influences of landscape and pollinators on population genetic structure: examples from three Penstemon (Plantaginaceae) species in the Great Basin. Amer J Bot 98:109–121. doi:10.3732/ajb.1000229
Li A, Ge S (2006) Genetic variation and conservation of Changnienia amoena, an endangered orchid endemic to China. Pl Syst Evol 258:251–260. doi:10.1007/s00606-006-0410-4
Lindley J (1836) Cattleya coccinea. Edwards’s Bot Reg 22:sub t.1919
Liu B, Wendel JF (2001) Intersimple sequence repeat (ISSR) polymorphisms as a genetic marker system in cotton. Molec Ecol Notes 1:205–208. doi:10.1046/j.1471-8278.2001.00073.x
Manuel R, Warren R, Miller D (1996) Sophronitis coccinea: a pollination study. Orchids 65:612–616
Menini Neto L, Barros F, Forzza RC (2007) Orchidaceae do Parque Estadual de Ibitipoca. Minas Gerais-Brasil. Acta Bot Brasil 21:687. doi:10.1590/S0102-33062007000300015
Micheneau C, Johnson SD, Fay MF (2009) Orchid pollination: from Darwin to the present day. Bot J Linn Soc 161:1–19. doi:10.1111/j.1095-8339.2009.00995.x
Miller D, Warren R, Miller I, Seehawer H (2006) Serra dos Órgãos, sua história, suas orquídeas. Gráfica Stamppa, Rio de Janeiro, pp 236–239
Miranda FG (1991) Sophronitis bicolor. Die Orchidee 42:227–230
Mort ME, Archibald JK, Randle CP, Levsen ND, O’Leary TR, Topalov K, Wiegand CM, Crawford DJ (2007) Inferring phylogeny at low taxonomic levels: utility of rapidly evolving cpDNA and nuclear ITS loci. Amer J Bot 94:173–183. doi:10.3732/ajb.94.2.173
Müller KF (2005) SeqState-primer design and sequence statistics for phylogenetic DNA datasets. Appl Bioinformatics 4:65–69. doi:10.2165/00822942-200504010-00008
Nei M (1973) Analysis of genetic diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Neiland MRM, Wilcock CC (1998) Fruit set, nectar reward, and rarity in the Orchidaceae. Amer J Bot 85:1657–1671
Nordström S, Hedrén M (2008) Genetic differentiation and postglacial migration of the Dactylorhiza majalis ssp. traunsteineri/lapponica complex into Fennoscandia. Pl Syst Evol 276:73–87. doi:10.1007/s00606-008-0084-1
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molec Ecol 13:1143–1155. doi:10.1111/j.1365-294X.2004.02141.x
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
Pabst GFJ (1976) Sophronitis coccinea subsp. pygmaea. Bradea 2(12):70
Pabst GFJ, Dungs F (1975) Orchidaceae brasiliensis I. Brücke-Verlag, Hildesheim
Pabst GFJ, Dungs F (1977) Orchidaceae brasiliensis II. Brücke-Verlag, Hildesheim
Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP (2010) Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol Evol 25:459–467. doi:10.1016/j.tree.2010.05.006
Phillips RD, Dixon KW, Peakall R (2012) Low population genetic differentiation in the Orchidaceae: implications for the diversification of the family. Molec Ecol 21:5208–5220. doi:10.1111/mec.12036
Pillon Y, Fay MF, Hedrén M, Bateman RM, Devey DS, Shipunov AB, van der Bank M, Chase MW (2007) Evolution and temporal diversification of western European polyploid species complexes in Dactylorhiza (Orchidaceae). Taxon 56:1185–1208. doi:10.2307/25065911
Pinheiro LR, Rabbani ARC, Silva AVC, Lédo AS, Pereira KLG, Leandro Diniz LEC (2012) Genetic diversity and population structure in the Brazilian Cattleya labiata (Orchidaceae) using RAPD and ISSR markers. Pl Syst Evol 298:1–11. doi:10.1007/s00606-012-0682-9
Pizo MA (2012) Lek behavior of the Plovercrest (Stephanoxis lalandi, Trochilidae). Wilson J Orni-thol 124:106–112. doi:10.1676/11-055.1
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rodriguez F, Oliver JL, Marin A, Medina JR (1990) The general stochastic model of nucleotide substitution. J Theor Biol 142:485–501
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi:10.1093/bioinformatics/btg180
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Molec Ecol Notes 4:137–138. doi:10.1046/j.1471-8286.2003.00566.x
Santos M (2001) Estradas reais: introdução ao estudo dos caminhos do ouro e dos diamantes no Brasil. Editora Estrada Real, Belo Horizonte
Scarcelli N, Barnaud A, Eiserhardt W, Treier UA, Seveno M, d’Anfray A, Pintaud JC (2011) A set of 100 chloroplast DNA primer pairs to study population genetics and phylogeny in monocotyledons. PLoS ONE. doi:10.1371/journal.pone.0019954
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Amer J Bot 92:142–166. doi:10.3732/ajb.92.1.142
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Amer J Bot 94:275–288. doi:10.3732/ajb.94.3.275
Silva-Pereira V, Smidt EDC, Borba EL (2007) Isolation mechanisms between two sympatric Sophronitis (Orchidaceae) species endemic to Northeastern Brazil. Pl Syst Evol 269:171–182. doi:10.1007/s00606-007-0583-5
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Smidt EC, Silva-Pereira V, Borba EL (2006) Reproductive biology of two Cattleya (Orchidaceae) species endemic to north-eastern Brazil. Pl Spec Biol 21:85–91. doi:10.1111/j.1442-1984.2006.00154.x
Sun M, Wong KC (2001) Genetic structure of three orchid species with contrasting breeding systems using RAPD and allozyme markers. Amer J Bot 88:2180–2188
Swofford DL (1998) PAUP: Phylogenetic analysis using parsimony (* and other methods), Version 4.0d64 ed. Sinauer Associates, Inc. Publishers, Sunderland
Swofford DL, Olsen GL (1990) Phylogeny Reconstruction. In: Hillis DM, Moritz C (eds) Molecular Systematics. Sinauer Associates Inc, Sunderland, pp 411–501
Szalanski AL, Steinauer G, Bischof R, Petersen J (2001) Origin and conservation genetics of the threatened Ute ladies’-tresses, Spiranthes diluvialis (Orchidaceae). Amer J Bot 88:177–180
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Pl Molec Biol 17:1105–1109
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Trends Ecol Evol 25:4876–4882
Tremblay RL (1992) Trends in pollination biology of the Orchidaceae: evolution and systematics. Can J Bot 70:642–650
Tremblay RL, Ackerman JD, Zimmerman JK, Calvo RN (2005) Variation in sexual reproduction in orchids and its evolutionary consequences: a spasmodic journey to diversification. Biol J Linn Soc 84:1–54. doi:10.1111/j.1095-8312.2004.00400.x
van den Berg C (2008) New combinations in the genus Cattleya (Orchidaceae). Neodiversity 3:3–12
van den Berg C, Chase MW (2000) Nomenclatural notes on Laeliinae–I. Lindleyana 15:115–119
van den Berg C, Higgins WE, Dressler RL, Whitten WM, Soto-Arenas MA, Culham A, Chase MW (2000) A phylogenetic analysis of Laeliinae (Orchidaceae) based on sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA. Lindleyana 15:96–114
van den Berg C, Higgins WE, Dressler RL, Whitten WM, Soto-Arenas MA, Chase MW (2009) A phylogenetic study of Laeliinae (Orchidaceae) based on combined nuclear and plastid DNA sequences. Ann Bot (Oxford) 104:417–430. doi:10.1093/aob/mcp101
van der Pijl L, Dodson CH (1966) Orchid flowers, their pollination and evolution. University of Miami Press, Coral Gables
Vieillot JP (1818) Nouveau dictionnaire d’histoire naturelle, appliquée aux arts, l’agriculture, à l’economie rurale et domestique, à la médicine, etc. Par une Société de Naturalistes et d’Agriculteurs, Paris
Whitten WM, Williams NH, Chase MW (2000) Subtribal and generic relationship of Maxillarieae (Orchidaceae) with emphasis on Stanhopeinae: combined molecular evidence. Amer J Bot 87:1842–1856
Withner CL (1993) The Cattleyas and their relatives: Schomburgkia, Sophronitis, and other South American genera. Timber Press, Portland
Wolfe AD (2000) ISSR protocols. http://www.biosci.ohio-state.edu/~awolfe/ISSR/protocols.ISSR.html. Accessed 18 May 2009
Wolfe AD, Liston A (1998) Contributions of PCR-based methods to plant systematics and evolutionary biology. In: Soltis DE, Soltis PS, Doyle JJ (eds) Plant molecular systematics II. Chapman Hall, New York, pp 43–86
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Alberta
Acknowledgments
We sincerely thank the “Instituto de Botânica de São Paulo” for providing conditions for cultivation of specimens for study, as well for all the help of their staff; D Miller, I Miller, LE Catharino, L Menini, MA Scalia, M Barros Neto, MR Cabral, O Ribeiro, R Valadares, T Campacci, W Forster for all the lab assistance and/or help on fieldwork; E Borba, F Pinheiro and two anonymous reviewers for contributions on earlier versions of this manuscript. Plants were collected under the permit SISBIO 32837-1. Funding was provided by Fundação de Amparo à Pesquisa do Estado de São Paulo/FAPESP (11/18751-7 to JFR, 11/18532-3 to EAV, 06/55121-3 to SK). CvdB and EAV thanks CNPq for a scholarship (Pq-1B and Pq-2).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Handling editor: Livia Wanntorp.
Rights and permissions
About this article
Cite this article
Rodrigues, J.F., van den Berg, C., Abreu, A.G. et al. Species delimitation of Cattleya coccinea and C. mantiqueirae (Orchidaceae): insights from phylogenetic and population genetics analyses. Plant Syst Evol 301, 1345–1359 (2015). https://doi.org/10.1007/s00606-014-1156-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-014-1156-z