Abstract
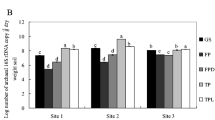
Drying soil samples before DNA extraction is commonly used for specific fungal DNA quantification and metabarcoding studies, but the impact of different drying procedures on both the specific fungal DNA quantity and the fungal community composition has not been analyzed. We tested three different drying procedures (freeze-drying, oven-drying, and room temperature) on 12 different soil samples to determine (a) the soil mycelium biomass of the ectomycorrhizal species Lactarius vinosus using qPCR with a specifically designed TaqMan® probe and (b) the fungal community composition and diversity using the PacBio® RS II sequencing platform. Mycelium biomass of L. vinosus was significantly greater in the freeze-dried soil samples than in samples dried at oven and room temperature. However, drying procedures had no effect on fungal community composition or on fungal diversity. In addition, there were no significant differences in the proportions of fungi according to their functional roles (moulds vs. mycorrhizal species) in response to drying procedures. Only six out of 1139 operational taxonomic units (OTUs) had increased their relative proportions after soil drying at room temperature, with five of these OTUs classified as mould or yeast species. However, the magnitude of these changes was small, with an overall increase in relative abundance of these OTUs of approximately 2 %. These results suggest that DNA degradation may occur especially after drying soil samples at room temperature, but affecting equally nearly all fungi and therefore causing no significant differences in diversity and community composition. Despite the minimal effects caused by the drying procedures at the fungal community composition, freeze-drying resulted in higher concentrations of L. vinosus DNA and prevented potential colonization from opportunistic species.


Similar content being viewed by others
References
Abarenkov K, Nilsson RH, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U (2010) The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol 186:281–285. doi:10.1111/j.1469-8137.2009.03160.x
Adams RI, Amend AS, Taylor JW, Bruns TD (2013a) A unique signal distorts the perception of species richness and composition in high-throughput sequencing surveys of microbial communities: a case study of fungi in indoor dust. Microb Ecol 66:735–741. doi:10.1007/s00248-013-0266-4
Adams RI, Miletto M, Taylor JW, Bruns TD (2013b) Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J 7:1262–1273. doi:10.1038/ismej.2013.28
Bååth E, Nilsson LO, Göransson H, Wallander H (2004) Can the extent of degradation of soil fungal mycelium during soil incubation be used to estimate ectomycorrhizal biomass in soil? Soil Biol Biochem 36:2105–2109. doi:10.1016/j.soilbio.2004.06.004
Bahram M, Peay KG, Tedersoo L (2014) Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytol 205:1454–1463. doi:10.1111/nph.13206
Bálint M, Bartha L, O’Hara RB, Olson MS, Otte J, Pfenninger M, Robertson A, Tiffin P, Schmitt I (2015) Relocation, high-latitude warming and host genetic identity shape the foliar fungal microbiome of poplars. Mol Ecol 24:235–248. doi:10.1111/mec.13018
Bonet JA, Martínez de Aragón JM, Pukkala T, Palahí M (2012) Immediate effect of thinning on the yield of Lactarius group deliciosus in Pinus pinaster forests in northeastern Spain. For Ecol Manag 265:211–217. doi:10.1016/j.foreco.2011.10.039
Cairney JWG (2005) Basidiomycete mycelia in forest soils: dimensions, dynamics and roles in nutrient distribution. Mycol Res 109:7–20. doi:10.1017/S0953756204001753
Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD (2015) Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol 205:1525–1536. doi:10.1111/nph.13208
Crecchio C, Stotzky G (1998) Binding of DNA on humic acids: effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol Biochem 30:1061–1067
De Caceres M, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. doi:10.1890/08-1823.1
De la Varga H, Águeda B, Ágreda T, Martínez-Peña F, Parladé J, Pera J (2013) Seasonal dynamics of Boletus edulis and Lactarius deliciosus extraradical mycelium in pine forests of central Spain. Mycorrhiza 23:391–402. doi:10.1007/s00572-013-0481-3
De Miguel S, Bonet JA, Pukkala T, Martínez de Aragón J (2014) Impact of forest management intensity on landscape-level mushroom productivity: a regional model-based scenario analysis. For Ecol Manag 330:218–227. doi:10.1016/j.foreco.2014.07.014
Feinstein LM, Sul WJ, Blackwood CB (2009) Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl Environ Microbiol 75:5428–5433. doi:10.1128/AEM.00120-09
Fernandez CW, McCormack ML, Hill JM, Pritchard SG, Koide RT (2013) On the persistence of Cenococcum geophilum ectomycorrhizas and its implications for forest carbon and nutrient cycles. Soil Biol Biochem 65:141–143. doi:10.1016/j.soilbio.2013.05.022
Filion M, St-arnaud M, Jabaji-hare SH (2003) Direct quantification of fungal DNA from soil substrate using real-time PCR. J Microbiol Methods 53:67–76
Gryndler M, Trilčová J, Hršelová H, Streiblová E, Gryndlerová H, Jansa J (2013) Tuber aestivum Vittad. mycelium quantified: advantages and limitations of a qPCR approach. Mycorrhiza 23:341–348. doi:10.1007/s00572-012-0475-6
Herdina NS, Jabaji-Hare S, Ophel-Keller K (2004) Persistence of DNA of Gaeumannomyces graminis var. tritici in soil as measured by a DNA-based assay. FEMS Microbiol Ecol 47:143–152. doi:10.1016/S0168-6496(03)00255-1
Hill MO (1973) Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432
Hortal S, Pera J, Parladé J (2008) Tracking mycorrhizas and extraradical mycelium of the edible fungus Lactarius deliciosus under field competition with Rhizopogon spp. Mycorrhiza 18:69–77. doi:10.1007/s00572-007-0160-3
Ihrmark K, Bodeker ITM, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandstrom- Durling M, Clemmensen KE, Lindahl BD (2012) New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 82:666–677. doi:10.1111/j.1574-6941.2012.01437.x
Karni M, Zidon D, Polak P, Zalevsky Z, Shefi O (2013) Thermal degradation of DNA. DNA Cell Biol 32:298–301. doi:10.1089/dna.2013.2056
Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Vrålstad T, Ursing BM (2005) UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068. doi:10.1111/j.1469-8137.2005.01376.x
Lindahl BD, Kuske CR (2013) Metagenomics for study of fungal ecology. In: Martin F (ed) The ecological genomics of fungi. John Wiley & Sons, Inc, Hoboken, NJ, pp. 281–303
Lindahl BD, de Boer W, Finlay RD (2010) Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J 4:872–881. doi:10.1038/ismej.2010.19
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J, Kåuserud H (2013) Fungal community analysis by high-throughput sequencing of amplified markers—a user’s guide. New Phytol 199:288–299. doi:10.1111/nph.12243
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi:10.1371/journal.pcbi.1003531
O’Hanlon R, Harrington TJ (2012) Similar taxonomic richness but different communities of ectomycorrhizas in native forests and non-native plantation forests. Mycorrhiza 22:371–382. doi:10.1007/s00572-011-0412-0
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHM, Wagner H (2015) VEGAN v2.1-10: community ecology package v 2.2-1 (http://cran.r-project.org/ web/packages/vegan/index.html, accessed 01.09.2015)
Parladé J, Hortal S, Pera J, Galipienso L (2007) Quantitative detection of Lactarius deliciosus extraradical soil mycelium by real-time PCR and its application in the study of fungal persistence and interspecific competition. J Biotechnol 128:14–23. doi:10.1016/j.jbiotec.2006.09.010
Parladé J, De la Varga H, De Miguel AM, Saéz R, Pera J (2013) Quantification of extraradical mycelium of Tuber melanosporum in soils from truffle orchards in northern Spain. Mycorrhiza 23:99–106. doi:10.1007/s00572-012-0454-y
Plassart P, Terrat S, Thomson B, Griffiths R, Dequiedt S, Lelievre M, Regnier T, Nowak V, Bailey M, Lemanceau P, Bispo A, Chabbi A, Maron PA, Mougel C, Ranjard L (2012) Evaluation of the ISO Standard 11063 DNA extraction procedure for assessing soil microbial abundance and community structure. PLoS One 7:1–8. doi:10.1371/journal.pone.0044279
R Core Team (2015) R: a language and environment for statistical computing. R Core Team, Vienna, Austria (http://cran.r-project.org/, accessed 09.10.2015)
Raidl S, Bonfigli R, Agerer R (2005) Calibration of quantitative real-time TaqMan PCR by correlation with hyphal biomass and ITS copies in mycelia of Piloderma croceum. Plant Biol 7:713–717. doi:10.1055/s-2005-873003
Roberts RJ, Carneiro MO, Schatz MC (2013) The advantages of SMRT sequencing. Genome Biol 14:405. doi:10.1186/gb-2013-14-6-405
Sterkenburg E, Bahr A, Brandström Durling M, Clemmensen KE (2015) Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol 207:1145–1158. doi:10.1111/nph.13426
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson K, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, Kesel AD, Dang T, Chen X, Buegger F, Brearley FQ (2014) Global diversity and geography of soil fungi. Science 346:1256688. doi:10.1126/science.1256688
Walkley A, Black A (1934) An examination of Degtjareff method for determining soil organic matter and a proposed modification of the chromic acid titration method. Soil Sci 37:29–37
Wallander H, Nilsson LO, Hagerberg D, Baath E (2001) Estimation of the biomass and seasonal growth of external mycelium of ectomycorrhizal fungi in the field. New Phytol 151:753–760. doi:10.1046/j.0028-646x.2001.00199
Wallander H, Ekblad A, Godbold DL, Johnson D, Bahr A, Baldrian P, Björk RG, Kieliszewska-Rokicka B, Kjøler R, Kraigher H, Plassard C, Rudawska M (2013) Evaluation of methods to estimate production, biomass and turnover of ectomycorrhizal mycelium in forests soils—a review. Soil Biol Biochem 57:1034–1047. doi:10.1016/j.soilbio.2012.08.027
Acknowledgments
The authors are very grateful to the PNIN of Poblet for its strong help in the installation and maintenance process of the experimental plots, and the Department of the Forest Mycology and Plant Pathology from SLU (Uppsala) for providing us the lab facilities. We are in debt with Björn Lindahl for the outstanding technical assistance in the DNA sequencing, and with Diem Nguyen for helping us with the statistical analysis of the fungal community. We are grateful for the critical review of the manuscript from Christine Fischer and the constructive comments and suggestions from two anonymous reviewers, which improved the quality of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interests.
Funding
This work was supported by a STSM Grant from COST Action FP1203 and by the Spanish Ministry of Economy and Competitivity (MINECO) through the project AGL 2012-40035-C03. Carles Castaño received support from the Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya through the program of Doctorats Industrials, funded by the European Union and the European Social Fund. Josu G. Alday was supported by Juan de la Cierva fellowships (IJCI-2014-21393).
Rights and permissions
About this article
Cite this article
Castaño, C., Parladé, J., Pera, J. et al. Soil drying procedure affects the DNA quantification of Lactarius vinosus but does not change the fungal community composition. Mycorrhiza 26, 799–808 (2016). https://doi.org/10.1007/s00572-016-0714-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-016-0714-3