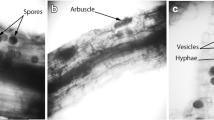
Abstract
An increasingly popular approach used to identify arbuscular mycorrhizal (AM) fungi in planta is to amplify a portion of AM fungal small subunit ribosomal DNA (SSU-rDNA) from whole root DNA extractions using the primer pair AM1-NS31, followed by cloning and sequencing. We used this approach to study the AM fungal community composition of three common oak-woodland plant species: a grass (Cynosurus echinatus), blue oak (Quercus douglasii), and a forb (Torilis arvensis). Significant diversity of AM fungi were found in the roots of C. echinatus, which is consistent with previous studies demonstrating a high degree of AM fungal diversity from the roots of various hosts. In contrast, clones from Q. douglasii and T. arvensis were primarily from non-AM fungi of diverse origins within the Ascomycota and Basidiomycota. This work demonstrates that caution must be taken when using this molecular approach to determine in planta AM fungal diversity if non-sequence based methods such as terminal restriction fragment length polymorphisms, denaturing gradient gel electrophoresis, or temperature gradient gel electrophoresis are used.


Similar content being viewed by others
References
Allen TR, Millar T, Berch SM, Berbee ML (2003) Culturing and direct DNA extraction find different fungi from the same ericoid mycorrhizal roots. New Phytol 160:255–272
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST—a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Clapp JP, Rodriguez A, Dodd (2002) Glomales rRNA gene diversity—all that glistens is not necessarily glomalean? Mycorrhiza 12:269–270
Dahlgren RA, Singer MJ, Huang X (1997) Oak tree and grazing impacts on soil properties and nutrients in a California oak woodland. Biogeochemistry 39:45–64
Daniell TJ, Husband R, Fitter AH, Young JPW (2001) Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microb Ecol 36:203–209
Douhan GW, Rizzo DM (2003) Amplified fragment length microsatellites (AFLM) might be used to develop microsatellite markers in organisms with limited amounts of DNA applied to arbuscular mycorrhizal (AM) fungi. Mycologia 95:368–373
Heijden MGA van der, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72
Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW (1998) Ploughing up the wood-wide Web? Nature 394:431
Helgason T, Fitter AH, Young JPW (1999) Molecular diversity of arbuscular mycorrhizal fungi colonising Hyacinthoides non-scripta (bluebell) in a seminatural woodland. Mol Ecol 8:659–666
Helgason T, Merryweather JW, Denison J, Wilson P, Young JPW, Fitter AH (2002) Selectivity and functional diversity in arbuscular mycorrhizas of co-occurring fungi and plants from a temperate deciduous woodland. J Ecol 90:371–384
Husband R, Herre EA, Turner SL, Gallery R, Young JPW (2002) Molecular diversity of arbuscular mycorrhizal fungi and patterns of host association over time and space in a tropical forest. Mol Ecol 11:2669–2678
Johnson D, Vandenkoornhuyse PJ, Leake JR, Gilbert L, Booth RE, Grime JP, Young JPW, Read DJ (2004) Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytol 161:503–515
Kjøller R, Rosendahl S (2000) Detection of arbuscular mycorrhizal fungi (Glomales) in roots by nested PCR and SSCP (single stranded conformation polymorphism). Plant Soil 226:189–196
Klironomos JN, McCune J, Hart M, Neville J (2000) The influence of arbuscular mycorrhizae on the relationship between plant diversity and productivity. Ecol Lett 3:137–141
Klironomos JN (2003) Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84:2292–2301
Kirk PM, Cannon PF, David JC, Stalpers JA (eds) (2001) Ainsworth and Bisby’s dictionary of the fungi, 9th edn. CABI, Egham, UK
Longato S, Bonfante P (1997) Molecular identification of mycorrhizal fungi by direct amplification of microsatellite regions. Mycol Res 101:425–432
Maddison DR, Maddison WP (2001) MacClade 4: analysis of phylogeny and character evolution, Version 4.03. Sinauer, Sunderland, Mass.
Millner PD, Mulbry WW, Reynolds SL (2001) Taxon-specific oligonucleotide primers for detection of Glomus etunicatum. Mycorrhiza 10:259–265
Morton JB, Redecker D (2001) Two new families of Glomales, Archaeosporaceae and Paraglomaceae, with two new genera Archaeospora and Paraglomus, based on concordant molecular and morphological characters. Mycologia 93:181–195
Qiu XY, Wu LY, Huang HS, McDonel PE, Palumbo AV, Tiedje JM, Zhou JZ (2001) Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl Environ Microbiol 67:880–887
Redecker D (2000) Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza 10:73–80
Redecker D, Morton JB, Bruns TD (2000) Ancestral lineages of arbuscular mycorrhizal fungi (Glomales). Mol Phylogenet Evol 14:276–284
Redecker D, Hijri I, Wiemken A (2003) Molecular identification of arbuscular mycorrhizal fungi in roots: perspectives and problems. Folia Geobot 38:113–124
Rosendahl S, Taylor JW (1997) Development of multiple genetic markers for studies of genetic variation in arbuscular mycorrhizal fungi using AFLP. Mol Ecol 6:821–829
Schwarzott D, Walker C, Schüßler A (2001) Glomus, the largest genus of the arbuscular mycorrhizal fungi (Glomales), is nonmonophyletic. Mol Phylogenet Evol 21:190–197
Schüßler A, Gehrig H, Schwarzott D, Walker C (2001a) Analysis of partial Glomales SSU rRNA gene sequences: implications for primer design and phylogeny. Mycol Res 105:5–15
Schüßler A, Schwarzott D, Walker C (2001b) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421
Simon L, Lalonde M, Bruns TD (1992) Specific amplification of the 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58:291–295
Smith SE, Read DJ (1993) Mycorrhizal symbiosis, 2nd edn. Academic, San Diego, Calif.
Swofford DL (2002) PAUP* phylogenetic analysis using parsimony (* and other methods), Version 4.0b10. Sinauer, Sunderland, Mass.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tuinen D van, Jacquot E, Zhao B, Gollotte A, Gianinazzi-Pearson V (1998) Characterization of root colonization profiles by a microcosm community of Arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol Ecol 7:879–887
Vandenkoornhuyse P, Husband R, Daniell TJ, Watson IJ, Duck JM, Fitter AH, Young JPW (2002) Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol Ecol 11:1555–1564
Vandenkoornhuyse P, Ridgway KP, Watson IJ, Fitter AH, Young JPW (2003) Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol Ecol 12:3085–3095
Wang GC, Wang Y (1996) The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107–1114
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Wintzingerode F von, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229
Wyss P, Bonfante P (1993) Amplification of genomic DNA of arbuscular-mycorrhizal (AM) fungi by PCR using short arbitrary primers. Mycol Res 97:1351–1357
Zézé A, Sulistyowati E, Ophel-Keller K, Barker S, Smith S (1997) Intersporal genetic variation of Gigaspora margarita, a vesicular arbuscular mycorrhizal fungus, revealed by M13 minisatellite-primed PCR. Appl Environ Microbiol 63:676–678
Acknowledgements
We thank Xinhua He for sharing unpublished root staining data, and two anonymous reviewers for helpful comments. This study was supported by the National Science Foundation Biocomplexity program (DEB 9981711, DEB 9981548)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Douhan, G.W., Petersen, C., Bledsoe, C.S. et al. Contrasting root associated fungi of three common oak-woodland plant species based on molecular identification: host specificity or non-specific amplification?. Mycorrhiza 15, 365–372 (2005). https://doi.org/10.1007/s00572-004-0341-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-004-0341-2