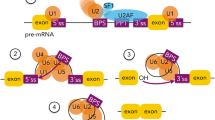
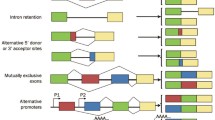
Abstract
Alternative splicing is a major cellular mechanism in metazoans for generating proteomic diversity. A large proportion of protein-coding genes in multicellular organisms undergo alternative splicing, and in humans, it has been estimated that nearly 90 % of protein-coding genes—much larger than expected—are subject to alternative splicing. Genomic analyses of alternative splicing have illuminated its universal role in shaping the evolution of genomes, in the control of developmental processes, and in the dynamic regulation of the transcriptome to influence phenotype. Disruption of the splicing machinery has been found to drive pathophysiology, and indeed reprogramming of aberrant splicing can provide novel approaches to the development of molecular therapy. This review focuses on the recent progress in our understanding of alternative splicing brought about by the unprecedented explosive growth of genomic data and highlights the relevance of human splicing variation on disease and therapy.

Similar content being viewed by others
References
Aaronson Y, Meshorer E (2013) Stem cells: regulation by alternative splicing. Nature 498:176–177
Aartsma-Rus A, van Ommen GJ (2009) Less is more: therapeutic exon skipping for Duchenne muscular dystrophy. Lancet Neurol 8:873–875
Aberg K, Saetre P, Jareborg N, Jazin E (2006) Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci USA 103:7482–7487
Alekseyenko AV, Kim N, Lee CJ (2007) Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA 13:661–670
Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK et al (2013) Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8:1765–1786
Asmann YW, Necela BM, Kalari KR, Hossain A, Baker TR et al (2012) Detection of redundant fusion transcripts as biomarkers or disease-specific therapeutic targets in breast cancer. Cancer Res 72:1921–1928
Baranauskas G, Tkatch T, Nagata K, Yeh JZ, Surmeier DJ (2003) Kv3.4 subunits enhance the repolarizing efficiency of Kv3.1 channels in fast-spiking neurons. Nat Neurosci 6:258–266
Barbosa-Morais NL, Carmo-Fonseca M, Aparicio S (2006) Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res 16:66–77
Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S et al (2012) The evolutionary landscape of alternative splicing in vertebrate species. Science 338:1587–1593
Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ et al (2013) The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. doi: 10.1038/mp.2013.45
Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR et al (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816
Brandenberger R, Wei H, Zhang S, Lei S, Murage J et al (2004) Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat Biotechnol 22:707–716
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571
Cavalier-Smith T (1991) Intron phylogeny: a new hypothesis. Trends Genet 7:145–148
Chandrasekharan NV, Dai H, Roos KL, Evanson NK, Tomsik J et al (2002) COX-3, a cyclooxygenase-1 variant inhibited by acetaminophen and other analgesic/antipyretic drugs: cloning, structure, and expression. Proc Natl Acad Sci USA 99:13926–13931
Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ et al (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5:613–619
Coulombe-Huntington J, Lam KC, Dias C, Majewski J (2009) Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet 5:e1000766
Crick F (1979) Split genes and RNA splicing. Science 204:264–271
Cusack BP, Wolfe KH (2005) Changes in alternative splicing of human and mouse genes are accompanied by faster evolution of constitutive exons. Mol Biol Evol 22:2198–2208
Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH (2008) The functional consequences of alternative promoter use in mammalian genomes. Trends Genet 24:167–177
DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD et al (2012) RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28:1530–1532
Dibb NJ (1991) Proto-splice site model of intron origin. J Theor Biol 151:405–416
Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK et al (2013) mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci 16:499–506
Dominski Z, Kole R (1993) Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci USA 90:8673–8677
Doolittle RF (1991) Counting and discounting the universe of exons. Science 253:677–680
Duan S, Huang RS, Zhang W, Mi S, Bleibel WK et al (2009) Expression and alternative splicing of folate pathway genes in HapMap lymphoblastoid cell lines. Pharmacogenomics 10:549–563
ENCODE Project Consortium (2004) The ENCODE (ENCyclopedia Of DNA Elements) project. Science 306:636–640
ENCODE Project Consortium (2011) A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9:e1001046
Ergun A, Doran G, Costello JC, Paik HH, Collins JJ et al (2013) Differential splicing across immune system lineages. Proc Natl Acad Sci USA 110:14324–14329
Fairbrother WG, Yeh RF, Sharp PA, Burge CB (2002) Predictive identification of exonic splicing enhancers in human genes. Science 297:1007–1013
Foissac S, Sammeth M (2007) ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35:W297–W299
Frankish A, Mudge JM, Thomas M, Harrow J (2012) The importance of identifying alternative splicing in vertebrate genome annotation. Database (Oxford) 2012:bas014
Gamazon ER, Duan S, Zhang W, Huang RS, Kistner EO et al (2010) PACdb: a database for cell-based pharmacogenomics. Pharmacogenet Genomics 20:269–273
Garcia-Blanco MA (2006) Alternative splicing: therapeutic target and tool. Prog Mol Subcell Biol 44:47–64
Gilbert W (1978) Why genes in pieces? Nature 271:501
GTEx Consortium (2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–585
Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B et al (2013) MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498:241–245
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M et al (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22:1760–1774
Hermiston ML, Xu Z, Majeti R, Weiss A (2002) Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J Clin Invest 109:9–14
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP et al (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106:9362–9367
Hogg M, Paro S, Keegan LP, O’Connell MA (2011) RNA editing by mammalian ADARs. Adv Genet 73:87–120
Hubbard T, Barker D, Birney E, Cameron G, Chen Y et al (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41
Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA et al (2002) A stem cell molecular signature. Science 298:601–604
Jacobsen M, Hoffmann S, Cepok S, Stei S, Ziegler A et al (2002) A novel mutation in PTPRC interferes with splicing and alters the structure of the human CD45 molecule. Immunogenetics 54:158–163
Jia G, Fu Y, He C (2013) Reversible RNA adenosine methylation in biological regulation. Trends Genet 29:108–115
Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM et al (2008) A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA 105:20333–20338
Katz Y, Wang ET, Airoldi EM, Burge CB (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7:1009–1015
Kim P, Kim N, Lee Y, Kim B, Shin Y et al (2005) ECgene: genome annotation for alternative splicing. Nucleic Acids Res 33:D75–D79
Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D et al (2009) Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol 8:918–928
Koo T, Wood MJ (2013) Clinical trials using antisense oligonucleotides in duchenne muscular dystrophy. Hum Gene Ther 24:479–488
Kwan T, Benovoy D, Dias C, Gurd S, Serre D et al (2007) Heritability of alternative splicing in the human genome. Genome Res 17:1210–1218
Kwan T, Benovoy D, Dias C, Gurd S, Provencher C et al (2008) Genome-wide analysis of transcript isoform variation in humans. Nat Genet 40:225–231
Lee Y, Gamazon ER, Rebman E, Lee S, Dolan ME et al (2012) Variants affecting exon skipping contribute to complex traits. PLoS Genet 8:e1002998
Li Q, Lee JA, Black DL (2007) Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci 8:819–831
Licatalosi DD, Darnell RB (2010) RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11:75–87
Lutz CS (2008) Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol 3:609–617
Martinez NM, Pan Q, Cole BS, Yarosh CA, Babcock GA et al (2012) Alternative splicing networks regulated by signaling in human T cells. RNA 18:1029–1040
Merkin J, Russell C, Chen P, Burge CB (2012) Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338:1593–1599
Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS et al (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 9:1415–1423
Modrek B, Lee CJ (2003) Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet 34:177–180
Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C et al (2010) Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464:773–777
Moroy T, Heyd F (2007) The impact of alternative splicing in vivo: mouse models show the way. RNA 13:1155–1171
Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN et al (2009) DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci USA 106:15873–15878
Pagani F, Buratti E, Stuani C, Romano M, Zuccato E et al (2000) Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J Biol Chem 275:21041–21047
Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P et al (2011) Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 365:1384–1395
Papasaikas P, Valcarcel J (2012) Evolution. Splicing in 4D. Science 338:1547–1548
Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE et al (2010a) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464:768–772
Pickrell JK, Pai AA, Gilad Y, Pritchard JK (2010b) Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet 6:e1001236
Planet E, Attolini CS, Reina O, Flores O, Rossell D (2012) htSeqTools: high-throughput sequencing quality control, processing and visualization in R. Bioinformatics 28:589–590
Pritsker M, Doniger TT, Kramer LC, Westcot SE, Lemischka IR (2005) Diversification of stem cell molecular repertoire by alternative splicing. Proc Natl Acad Sci USA 102:14290–14295
Pruitt KD, Maglott DR (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res 29:137–140
Qiu WG, Schisler N, Stoltzfus A (2004) The evolutionary gain of spliceosomal introns: sequence and phase preferences. Mol Biol Evol 21:1252–1263
Roberts JM, Ennajdaoui H, Edmondson C, Wirth B, Sanford J et al (2013) Splicing factor TRA2B is required for neural progenitor survival. J Comp Neurol 522(2):372–392
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E et al (2008) Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res 18:88–103
Schwartz S, Meshorer E, Ast G (2009) Chromatin organization marks exon–intron structure. Nat Struct Mol Biol 16:990–995
Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33:787–791
St Clair D, Blackwood D, Muir W, Carothers A, Walker M et al (1990) Association within a family of a balanced autosomal translocation with major mental illness. Lancet 336:13–16
Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S et al (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1:e78
Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP et al (2007) Population genomics of human gene expression. Nat Genet 39:1217–1224
Sugnet CW, Kent WJ, Ares M Jr, Haussler D (2004) Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput 2004:66–77
Thorn CF, Klein TE, Altman RB (2005) PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol 311:179–191
Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M et al (2009) Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16:996–1001
Trapnell C, Roberts A, Goff L, Pertea G, Kim D et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Vandiedonck C, Taylor MS, Lockstone HE, Plant K, Taylor JM et al (2011) Pervasive haplotypic variation in the spliceo-transcriptome of the human major histocompatibility complex. Genome Res 21:1042–1054
Wang Z, Burge CB (2008) Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14:802–813
Wang Z, Rolish ME, Yeo G, Tung V, Mawson M et al (2004) Systematic identification and analysis of exonic splicing silencers. Cell 119:831–845
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L et al (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang Y, Ma M, Xiao X, Wang Z (2012) Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol 19:1044–1052
Wong MS, Chen L, Foster C, Kainthla R, Shay JW et al (2013) Regulation of telomerase alternative splicing: a target for chemotherapy. Cell Rep 3:1028–1035
Xing Y, Lee C (2005) Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc Natl Acad Sci USA 102:13526–13531
Xu X, Yang D, Ding JH, Wang W, Chu PH et al (2005) ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell 120:59–72
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Gamazon, E.R., Stranger, B.E. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet 133, 679–687 (2014). https://doi.org/10.1007/s00439-013-1411-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-013-1411-3