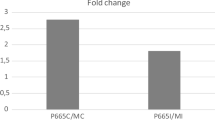
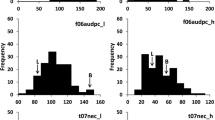
Abstract
Key message
Phenotyping of specific cellular resistance responses and improvement of previous genetic map allowed the identification of novel genomic regions controlling cellular mechanisms involved in pea resistance to ascochyta blight and provided candidate genes suitable for MAS.
Abstract
Didymella pinodes, causing ascochyta blight, is a major pathogen of the pea crop and is responsible for serious damage and yield losses. Resistance is inherited polygenically and several quantitative trait loci (QTLs) have been already identified. However, the position of these QTLs should be further refined to identify molecular markers more closely linked to the resistance genes. In previous works, resistance was scored visually estimating the final disease symptoms; in this study, we have conducted a more precise phenotyping of resistance evaluating specific cellular resistance responses at the histological level to perform a more accurate QTL analysis. In addition, P665 × Messire genetic map used to identify the QTLs was improved by adding 117 SNP markers located in genes. This combined approach has allowed the identification, for the first time, of genomic regions controlling cellular mechanisms directly involved in pea resistance to ascochyta blight. Furthermore, the inclusion of the gene-based SNP markers has allowed the identification of candidate genes co-located with QTLs and has provided robust markers for marker-assisted selection.



Similar content being viewed by others
References
Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111:9
Ascencio-Ibáñez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148:436–454
Aubert G, Morin J, Jacquin F, Loridon K, Quillet MC, Petit A, Rameau C, Lejeune-Hénaut I, Huguet T, Burstin J (2006) Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor Appl Genet 112:1024–1041
Aubry S, Mani J, Hörtensteiner S (2008) Stay-green protein, defective Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide an oxygenase in the chlorophyll catabolic pathway. Plant Mol Biol 67:243–256
Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488
Boisson B, Giglione C, Meinnel T (2003) Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J Biol Chem 278:43418–43429
Bordat A, Savois V, Nicolas M, Salse J, Chauveau A, Bourgeois M, Burstin J (2011) Translational genomics in legumes allowed placing in silico 5460 unigenes on the pea functional map and identified candidate genes in Pisum sativum L. G3 Gene Genome Genet 1:93–103
Borisov AY, Madsen LH, Tsyganov E, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, Tikhonovich IA, Stougaard J (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicas. Plant Physiol 131:1009–1017
Bretag TW (1989) Resistance of pea cultivars to ascochyta blight caused by Mycosphaerella pinodes, Phoma medicaginis and Ascochyta pisi. Ann Appl Biol 114:156–157
Bretag TW (1991) Epidemiology and control of ascochyta blight of field peas. PhD Thesis, La Trobe University, Australia
Bretag TW, Keane PJ, Price TV (2006) The epidemiology and control of ascochyta blight in field peas: a review. Aust J Agric Res 57:883–902
Carrillo E, Rubiales D, Pérez-de-Luque A, Fondevilla S (2013) Characterization of mechanisms of resistance against Didymella pinodes in Pisum spp. Eur J Plant Pathol 135:761–769
Chang MM, Horovitz D, Culley D, Hadwiger LA (1995) Molecular cloning and characterization of a pea chitinase gene expressed in response to wounding, fungal infection and the elicitor chitosan. Plant Mol Biol 28:105–111
Choi HK, Luckow MA, Doyle J, Cook DR (2006) Development of nuclear gene-derived molecular markers linked to legume genetics maps. Mol Genet Genom 276:56–70
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Clark VL (2004) SAS/STAT 91: User’s guide. SAS Institute Inc, Cary
Clulow SA, Lewis BG, Matthews P (1991) A pathotype classification for Mycosphaerella pinodes. J Phytopathol 131:322–332
Darvasi A, Soller M (1997) A simple method to calculate resolving power and confidence interval of QTL map location. Behav Genet 27:125–132
DeMason DA, Weeden NF (2006) Two Argonaute 1 genes from pea. Pisum Genet 38:3–9
Deulvot C, Charrel H, Marty A, Jacquin F, Donnadieu C, Lejeune-Hénaut I, Aubert G (2010) Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genom 11:468
Edwards A, Heckmann AB, Yousafzai F, Duc G, Downie JA (2007) Structural implications of mutation in the pea SYM8 symbiosis gene, the DMIl ortholog, encoding a predicted ion cannel. Mol Plant Microbe Interact 20:1183–1191
FAOSTAT (2012) Statistical databases. Food and Agriculture Organization of the United Nations
Fernández-Aparicio M, Amri M, Kharrat M, Rubiales D (2010) Intercropping reduces Mycosphaerella pinodes severity and delays upward progress on the pea plant. Crop Prot 29:744–750
Fondevilla S, Ávila CM, Cubero JI, Rubiales D (2005) Response to Mycosphaerella pinodes in a germplasm collection of Pisum spp. Plant Breed 124:313–315
Fondevilla S, Cubero JI, Rubiales D (2007) Inheritance of resistance to Mycosphaerella pinodes in two wild accessions of Pisum. In: Tivoli B, Baranger A, Muehlbauer FJ, Cooke BM (eds) Ascochyta blights of grain legumes. Springer, Netherlands, pp 53–58
Fondevilla S, Satovic Z, Rubiales D, Moreno MT, Torres AM (2008) Mapping of quantitative trait loci for resistance to Mycosphaerella pinodes in Pisum sativum subsp syriacum. Mol Breed 21:439–454
Fondevilla S, Almeida NF, Satovic Z, Rubiales D, Patto MCV, Cubero JI, Torres AM (2011) Identification of common genomic regions controlling resistance to Mycosphaerella pinodes, earliness and architectural traits in different pea genetic backgrounds. Euphytica 182:43–52
Fondevilla S, Martín-Sanz A, Satovic Z, Fernández-Romero MD, Rubiales D, Caminero C (2012) Identification of quantitative trait loci involved in resistance to Pseudomonas syringae pv syringae in pea (Pisum sativum L.). Euphytica 186:805–812
Fondevilla S, Rotter B, Krezdorn N, Jüngling R, Winter P, Rubiales D (2013) Identification of genes involved in resistance to Didymella pinodes in pea by deepSuperSAGE genome-wide transcriptome profiling. Book of abstracts of First Legume Society Conference, pp 148
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131:1855–1867
Jing R, Johnson R, Seres A, Kiss G, Ambrose MJ, Knox MR, Ellis THN, Flavell AJ (2007) Gene-based sequence diversity analysis of field pea (Pisum). Genetics 177:2263–2275
Kao CH, Zeng ZB, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216
Khan TN, Timmerman-Vaughan GM, Rubiales D, Warkentin TD, Siddique KHM, Erskine W, Barbetti MJ (2013) Didymella pinodes and its management in field pea: challenges and opportunities. Field Crop Res 148:61–77
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kraft JM, Dunne B, Goulden D, Armstrong S (1998) A search for resistance in peas to Mycosphaerella pinodes. Plant Dis 82:251–253
Krussell L, Sato N, Fukuhara I, Koch BE, Grossmann C, Okamoto S, Oka-Kira E, Otsubo Y, Aubert G, Nakagawa T, Sato S, Tabata S, Duc G, Parniske M, Wang TL, Kawaguchi M, Stougaard J (2011) The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J 65:861–871
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lawyer AS (1984) Diseases caused by Ascochyta spp. In: Hagedorn DJ (ed) The compendium of pea diseases. American Phytopathological Society, Wisconsin, pp 11–15
Marrs KA (1996) The functions and regulation of glutathione S-transferases in plants. Ann Rev Plant Biol 47:127–158
McMurray LS, Davidson JA, Lines MD, Leonforte A, Salam MU (2011) Combining management and breeding advances to improve field pea (Pisum sativum L.) grain yields under changing climatic conditions in south-eastern Australia. Euphytica 180:69–88
Mellersh DG, Foulds IV, Higgins VJ, Heath MC (2002) H2O2 plays different roles in determining penetration failure in three diverse plant–fungal interactions. Plant J 29:257–268
Nasir M, Hoppe HH (1997) Evaluation of varietal susceptibility in peas to Mycosphaerella pinodes. Test Agrochem Cv 18:32–33
Prioul S, Onfroy C, Tivoli B, Baranger A (2003) Controlled environment assessment of partial resistance to Mycosphaerella pinodes in pea (Pisum sativum L.) seedlings. Euphytica 131:121–130
Prioul S, Frankewitz A, Deniot G, Morin G, Baranger A (2004) Mapping of quantitative trait loci for partial resistance to Mycosphaerella pinodes in pea (Pisum sativum L.), at the seedling and adult plant stages. Theor Appl Genet 108:1322–1334
Prioul-Gervais S, Deniot G, Receveur EM, Frankewitz A, Fourmann M, Rameau C, Baranger A (2007) Candidate genes for quantitative resistance to Mycosphaerella pinodes in pea (Pisum sativum L.). Theor Appl Genet 114:971–984
Qi Y, Katagiri F (2012) Membrane microdomain may be a platform for immune signaling. Plant Signal Behav 7:454–456
Razdan K, Heinrikson RL, Zurcher-Neely H, Morris PW, Anderson LE (1992) Chloroplast and cytoplasmic enzymes: isolation and sequencing of cDNAs coding for two distintic pea chloroplast aldolases. Arch Biochem Biophys 298:192–197
Renner T, Specht C (2012) Molecular and functional evolution of class I chitinases for plant carnivory in the Caryophyllales. Mol Biol Evol 29:2971–2985
Roger C, Tivoli B (1996) Spatio-temporal development of pycnidia and perithecia and dissemination of spores of Mycosphaerella pinodes on pea (Pisum sativum). Plant Pathol 45:518–528
Rubiales D, Fondevilla S (2012) Future prospects for ascochyta blight resistance breeding in cool season food legumes. Front Plant Sci 3:27
Rubiales D, Pérez-de-Luque A, Cubero JI, Sillero JC (2003) Crenate broomrape (Orobanche crenata) infection in field pea cultivars. Crop Prot 22:865–872
Rubiales D, Fernández-Aparicio M, Moral A, Barilli E, Sillero JC, Fondevilla S (2009) Disease resistance in pea (Pisum sativum L.) types for autumn sowings in Mediterranean environments. Czech J Genet Plant Breed 45:135–142
Schoeny A, Menat J, Darsonval A, Rouault F, Jumel S, Tivoli B (2008) Effect of pea canopy architecture on splash dispersal of Mycosphaerella pinodes conidia. Plant Pathol 57:1073–1085
Seki H, Nakamura N, Marutani M, Okabe T, Sanematsu S, Inagaki Y, Ichinose Y (2002) Molecular cloning of cDNA for a novel pea Dof protein, PsDof1, and its DNA-binding activity to the promoter of PsDof1 gene. Plant Biotechnol 19:251–260
Stuckey HP (1940) Georgia experiment station annual report. In: University System of Georgia, U (ed) Georgia Experiment Station, Georgia, USA, pp 50–64
Tar’an B, Warkentin T, Somers DJ, Miranda D, Vandenberg A, Blade S, Penner G (2003) Quantitative trait loci for lodging resistance, plant height and partial resistance to Mycosphaerella blight in field pea (Pisum sativum L.). Theor Appl Genet 107:1482–1491
Timmerman-Vaughan GM, Frew TJ, Russell AC, Khan T, Butler R, Gilpin M, Falloon K (2002) QTL mapping of partial resistance to field epidemics of ascochyta blight of pea. Crop Sci 42:2100–2111
Timmerman-Vaughan GM, Frew TJ, Butler R, Murray S, Gilpin M, Falloon K, Khan T (2004) Validation of quantitative trait loci for Ascochyta blight resistance in pea (Pisum sativum L.) using populations from two crosses. Theor Appl Genet 109:1620–1631
Tivoli B, Banniza S (2007) Comparison of the epidemiology of ascochyta blights on grain legumes. In: Tivoli B, Baranger A, Muehlbauer FJ, Cooke BM (eds) Ascochyta blights of grain legumes. Springer, Netherlands, pp 59–76
Tivoli B, Béasse C, Lemarchand E, Masson E (1996) Effect of ascochyta blight (Mycosphaerella pinodes) on yield components of single pea (Pisum sativum) plants under field conditions. Ann Appl Biol 129:207–216
Tivoli B, Baranger A, Avila CM, Banniza S, Barbetti M, Chen W, Davidson J, Lindeck K, Kharrat M, Rubiales D, Sadiki M, Sillero JC, Sweetingham M, Muehlbauer FJ (2006) Screening techniques and sources of resistance to foliar diseases caused by major necrotrophic fungi in grain legumes. Euphytica 147:223–253
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell Online 15:533–543
Wallen VR (1965) Field evaluation and the importance of the Ascochyta complex on peas. Can J Plant Sci 45:27–33
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL cartographer 25 Department of Statistics. North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/QTLcart/WQTLCarthtm
Weller JL, Liew LC, Hecht VF, Rajandran V, Laurie RE, Ridge S, Lejeune-Hénaut I (2012) A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Nat Acad Sci 109:21158–21163
Wroth JM (1996) Host-pathogen relationship of the ascochyta blight (Mycosphaerella pinodes (Berk Blox) Vesterg) disease of field pea (Pisum sativum L.). PhD thesis University of Western Australia, Perth
Wroth JM (1998) Possible role for wild genotypes of Pisum spp to enhance ascochyta blight resistance in pea. Anim Prod Sci 38:469–479
Wroth JM (1999) Evidence suggests that Mycosphaerella pinodes infection of Pisum sativum is inherited as a quantitative trait. Euphytica 107:193–204
Wroth JM, Khan TN (1999) Differential responses of field pea (Pisum sativum L.) to ascochyta blight (Mycosphaerella pinodes): rating disease in the field. Aus J Agric Res 50:601–615
Wu Y, Bhat PR, Close TJ, Lonardi S (2008) Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet 4:e1000212
Xue AG, Warkentin TD (2001) Partial resistance to Mycosphaerella pinodes in field pea. Can J Plant Sci 81:535–540
Zeng ZB, Kao CH, Basten CJ (1999) Estimating the genetic architecture of quantitative traits. Genet Res 74:279–289
Zhang R, Hwang SF, Chang KF, Gossen BD, Strelkov SE, Turnbull GD, Blade SF (2006) Genetic resistance to in 558 field pea accessions. Crop Sci 46:2409–2414
Acknowledgments
E. Carrillo was granted by a Cabildo de La Palma-CSIC PhD grant and S. Fondevilla by a MC contract: FP7-PEOPLE-2011-IEF-300235. Financial support by projects AGL2011-22524 (co-financed by FEDER) and FP7-ARIMNet-Medileg is acknowledged. The authors wish to thank Sophie Valière and Cécile Donnadieu for the use of the GeT-PlaGe Genotyping platform and their expertise.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments comply with the current laws of the country in which they were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Puigdomenech.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carrillo, E., Satovic, Z., Aubert, G. et al. Identification of quantitative trait loci and candidate genes for specific cellular resistance responses against Didymella pinodes in pea. Plant Cell Rep 33, 1133–1145 (2014). https://doi.org/10.1007/s00299-014-1603-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1603-x