Abstract
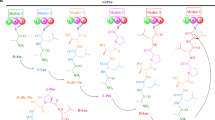
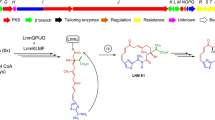
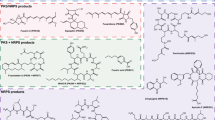
Sansalvamide is a cyclic pentadepsipeptide produced by Fusarium solani and has shown promising results as potential anti-cancer drug. The biosynthetic pathway has until now remained unidentified, but here we used an Agrobacterium tumefaciens-mediated transformation (ATMT) approach to generate knockout mutants of two candidate non-ribosomal peptide synthetases (NRPS29 and NRPS30). Comparative studies of secondary metabolites in the two deletion mutants and wild type confirmed the absence of sansalvamide in the NRPS30 deletion mutant, implicating this synthetase in the biosynthetic pathway for sansalvamide. Sansalvamide is structurally related to the cyclic hexadepsipeptide destruxin, which both contain an α-hydroxyisocaproic acid (HICA) unit. A gene cluster responsible for destruxin production has previously been identified in Metarhizium robertsii together with a hypothetical biosynthetic pathway. Using comparative bioinformatic analyses of the catalytic domains in the destruxin and sansalvamide NRPSs, we were able to propose a model for sansalvamide biosynthesis. Orthologues of the gene clusters were also identified in species from several other genera including Acremonium chrysogenum and Trichoderma virens, which suggests that the ability to produce compounds related to destruxin and sansalvamide is widespread.





Similar content being viewed by others
References
Amiri-Besheli B, Khambay B, Cameron S, Deadman ML, Butt TM (2000) Inter- and intra-specific variation in destruxin production by insect pathogenic Metarhizium spp., and its significance to pathogenesis. Mycol Res 104:447–452
Belofsky GN, Jensen PR, Fenical W (1999) Sansalvamide: a new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett 40:2913–2916
Boc A, Diallo AB, Makarenkov V (2012) T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res 40:W573–W579
Challis GL, Naismith JH (2004) Structural aspects of non-ribosomal peptide biosynthesis. Curr Opin Struct Biol 14:748–756
Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, Grimwood J, Schmutz J, Taga M, White GJ, Zhou S, Schwartz DC, Freitag M, L-j Ma, Danchin EGJ, Henrissat B, Coutinho PM, Nelson DR, Straney D, Napoli CA, Barker BM, Gribskov M, Rep M, Kroken S, Molnar I, Rensing C, Kennell JC, Zamora J, Farman ML, Selker EU, Salamov A, Shapiro H, Pangilinan J, Lindquist E, Lamers C, Grigoriev IV, Geiser DM, Covert SF, Temporini E, VanEtten HD (2009) The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet 5:e1000618
Coleman JJ, White GJ, Rodriguez-Carres M, VanEtten HD (2011) An ABC transporter and a cytochrome P450 of Nectria haematococca MPVI are virulence factors on pea and are the major tolerance mechanisms to the Phytoalexin pisatin. Mol Plant Microbe Interact 24:368–376
Cueto M, Jensen PR, Fenical W (2000) N-methylsansalvamide, a cytotoxic cyclic depsipeptide from a marine fungus of the genus Fusarium. Phytochemistry 55:223–226
de Groot MJA, Bundock P, Hooykaas PJJ, Beijersbergen AGM (1998) Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol 16:839–842
Dickman MB, Kolattukudy PE (1987) Transformation of Fusarium solani pisi using the Cutinase Promoter. Phytopathology 77:1740
Donzelli BGG, Krasnoff SB, Sun-Moon Y, Churchill ACL, Gibson DM (2012) Genetic basis of destruxin production in the entomopathogen Metarhizium robertsii. Curr Genet 58:105–116
Edel-Hermann V, Gautheron N, Mounier A, Steinberg C (2015) Fusarium diversity in soil using a specific molecular approach and a cultural approach. J Microbiol Methods 111:64–71
Frandsen RJN, Nielsen NJ, Maolanon N, Sørensen JC, Olsson S, Nielsen J, Giese H (2006) The biosynthetic pathway for aurofusarin in Fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol Microbiol 61:1069–1080
Frandsen RJN, Andersson JA, Kristensen MB, Giese H (2008) Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol Biol 9:70
Fullner KJ, Nester EW (1996) Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J Bacteriol 178:1498–1504
Graziani S, Vasnier C, Daboussi MJ (2004) Novel polyketide synthase from Nectria haematococca. Appl Environ Microbiol 70:2984–2988
Hansen FT, Droce A, Sørensen JL, Fojan P, Giese H, Sondergaard TE (2012a) Overexpression of NRPS4 leads to increased surface hydrophobicity in Fusarium graminearum. Fungal Biol 116:855–862
Hansen FT, Sørensen JL, Giese H, Sondergaard TE, Frandsen RJN (2012b) Quick guide to polyketide synthase and nonribosomal synthetase genes in Fusarium. Int J Food Microbiol 155:128–136
Hansen FT, Gardiner DM, Lysoe E, Fuertes PR, Tudzynski B, Wiemann P, Sondergaard TE, Giese H, Brodersen DE, Sørensen JL (2015) An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet Biol 75:20–29
Helaers R, Milinkovitch MC (2010) MetaPIGA v2.0: maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics 11:379
Hwang Y, Rowley D, Rhodes D, Gertsch J, Fenical W, Bushman F (1999) Mechanism of inhibition of a poxvirus topoisomerase by the marine natural product sansalvamide A. Mol Pharmacol 55:1049–1053
Jin SG, Komari T, Gordon MP, Nester EW (1987) Genes responsible for the supervirulence phenotype of Agrobacterium tumefaciens a281. J Bacteriol 169:4417–4425
Khayatt BI, Overmars L, Siezen RJ, Francke C (2013) Classification of the adenylation and acyl-transferase activity of NRPS and PKS systems using ensembles of substrate specific hidden Markov models. PLoS One 8:e62136
Lee H-S, Lee C (2012) Structural analysis of a new cytotoxic demethylated analogue of neo-n-methylsansalvamide with a different peptide sequence produced by Fusarium solani isolated from potato. J Agr Food Chem 60:4342–4347
Lee H-S, Phat C, Nam WS, Lee C (2014) Optimization of culture conditions of Fusarium solani for the production of neoN-methylsansalvamide. Biosci Biotechnol Biochem 78:1421–1427
Liu H, Bao X (2009) Overexpression of the chitosanase gene in Fusarium solani via Agrobacterium tumefaciens-mediated transformation. Curr Microbiol 58:279–282
Liu B-L, Tzeng Y-M (2012) Development and applications of destruxins: a review. Biotechnol Adv 30:1242–1254
Malz S, Grell MN, Thrane C, Maier FJ, Rosager P, Felk A, Albertsen KS, Salomon S, Bohn L, Schafer W, Giese H (2005) Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet Biol 42:420–433
Nalim FA, Samuels GJ, Wijesundera RL, Geiser DM (2011) New species from the Fusarium solani species complex derived from perithecia and soil in the Old World tropics. Mycologia 103:1302–1330
Nucci M, Anaissie E (2007) Fusarium infections in immunocompromised patients. Clin Microbiol Rev 20:695–704
O’Donnell K (2000) Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919–938
O’Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, Zhang N, Geiser DM (2008) Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol 46:2477–2490
Sandrock RW, Vanetten HD (2001) The relevance of tomatinase activity in pathogens of tomato: disruption of the beta(2)-tomatinase gene in Colletotrichum coccodes and Septoria lycopersici and heterologous expression of the Septoria lycopersici beta(2)-tomatinase in Nectria haematococca, a pathogen of tomato fruit. Physiol Mol Plant Pathol 58:159–171
Short DPG, O’Donnell K, Thrane U, Nielsen KF, Zhang N, Juba JH, Geiser DM (2013) Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp nov and F. petroliphilum stat. nov. Fungal Genet Biol 53:59–70
Simmons EG (1992) Alternaria: Biology, plant diseases and metabolites. In: Chelkowski J, Visconti A (eds) Alternaria taxonomy: current status, viewpoints, challenge. Elsevier, Amsterdam, pp 1–36
Sivashanmugam M, Nagarajan H, Vetrivel U, Ramasubban G, Therese KL, Narahari MH (2015) In silico analysis and prioritization of drug targets in Fusarium solani. Med Hypotheses 84:81–84
Slightom JL, Metzger BR, Luu HT, Elhammer AP (2009) Cloning and molecular characterization of the gene encoding the Aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 431:67–79
Smedsgaard J (1997) Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J Chromatogr A 760:264–270
Song H-H, Lee H-S, Lee C (2011) A new cytotoxic cyclic pentadepsipeptide, neo-N-methylsansalvamide produced by Fusarium solani KCCM90040, isolated from potato. Food Chem 126:472–478
Sørensen JL, Sondergaard TE (2014) The effects of different yeast extracts on secondary metabolite production in Fusarium. Int J Food Microbiol 170:55–60
Srivastava SK, Huang X, Brar HK, Fakhoury AM, Bluhm BH, Bhattacharyya MK (2014) The genome sequence of the fungal pathogen Fusarium virguliforme that cause sudden death syndrome in soybean. Plos One 9:e81832
Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6:493–505
Strieker M, Tanovic A, Marahiel MA (2010) Nonribosomal peptide synthetases: structures and dynamics. Curr Opin Struct Biol 20:234–240
Uhlig S, Jestoi M, Knutsen AK, Heier BT (2006) Multiple regression analysis as a tool for the identification of relations between semi-quantitative LC-MS data and cytotoxicity of extracts of the fungus Fusarium avenaceum (syn. F-arthrosporioides). Toxicon 48:567–579
Wang B, Kang Q, Lu Y, Bai L, Wang C (2012) Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Natl Acad Sci USA 109:1287–1292
Wasmann CC, VanEtten HD (1996) Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol Plant Microbe Interact 9:793–803
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Mueller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243
Yoder WT, Christianson LM (1998) Species-specific primers resolve members of Fusarium section Fusarium—Taxonomic status of the edible “Quorn” fungus reevaluated. Fungal Genet Biol 23:68–80
Zaccardelli M, Vitale S, Luongo L, Merighi M, Corazza L (2008) Morphological and molecular characterization of Fusarium solani isolates. J Phytopathol 156:534–541
Zhang N, O’Donnell K, Sutton DA, Nalim FA, Summerbell RC, Padhye AA, Geiser DM (2006) Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol 44:2186–2190
Zhang H, Gao S, Lercher MJ, Hu S, Chen WH (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40:W569–W572
Acknowledgments
Agrobacterium tumefaciens AGL-1 was a kind gift from Professor Birgitte K. Ahring and PhD students Lei Yang and Istvan Weyda at Aalborg University. Funding for this project was provided by the Aarhus University Research Foundation (AUFF) NANORIPES Centre for Natural Non-Ribosomal Peptide Synthesis (DEB).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Romans-Fuertes, P., Sondergaard, T.E., Sandmann, M.I.H. et al. Identification of the non-ribosomal peptide synthetase responsible for biosynthesis of the potential anti-cancer drug sansalvamide in Fusarium solani . Curr Genet 62, 799–807 (2016). https://doi.org/10.1007/s00294-016-0584-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0584-4