Abstract
In the present study, we characterize a polymorphism in the CD93 molecule, originally identified as the receptor for the C1q complement component (i.e., C1qRp, or AA4.1) in non-obese diabetic (NOD) mice. This allele carries a coding polymorphism in the first epidermal growth factor-like domain of CD93, which results in an amino acid substitution from Asn→His at position 264. This polymorphism does not appear to influence protein translation or ecto-domain cleavage, as CD93 is detectable in bone-marrow-derived macrophage and B-cell precursor lysates and in soluble form in the serum. The NOD CD93 isoform causes a phenotypic aberrancy in the early B-cell developmental stages (i.e., pro-, pre-, immature, and transitional), likely related to a conformational variation. Interestingly, the NZB/W F1 strain, which serves as a murine model of Lupus, also expresses an identical CD93 sequence polymorphism. Cd93 is located within the NOD Idd13 locus and is also tightly linked to the NZB/W F1 Wbw1 and Nkt2 disease susceptibility loci, which are thought to regulate natural killer T (NKT) cell homeostasis. Consistent with this genetic linkage, we found B6 CD93−/− and B6.NODIdd13 mice to be susceptible to a profound CD4+ NKT cell deficient state. These data suggest that Cd93 may be an autoimmune susceptibility gene residing within the Idd13 locus, which plays a role in regulating absolute numbers of CD4+ NKT cells.
Similar content being viewed by others
Introduction
CD93 is expressed by endothelial cells, cells of myeloid lineage, platelets, and early hematopoietic stem cells and is an important lineage-specific marker of early B-cell developmental stages (Cancro 2004; Danet et al. 2002; Fonseca et al. 2001; Nepomuceno et al. 1997; Nepomuceno and Tenner 1998; Petrenko et al. 1999). Normally, CD93 is expressed at high levels on pro-, pre- and immature bone marrow (BM) B-cell progenitors as well as transitional (TR) B cells in the periphery (Cancro 2004). The Cd93 gene is located at 84 cM on murine chromosome 2 (Kim et al. 2000) and encodes a type I O-glycosylated transmembrane protein whose domain structure includes an amino-terminal C-type lectin domain, a tandem array of five epidermal growth factor (EGF)-like repeats, a single hydrophobic transmembrane region, and a short cytoplasmic domain that contains a PDZ binding domain and a moesin interaction site (Bohlson et al. 2005; Kim et al. 2000; Norsworthy et al. 1999; Petrenko et al. 1999; Zhang et al. 2005). This domain structure bears a unique resemblance to the selectin family of adhesion molecules (Dean et al. 2001; Kim et al. 2000; Norsworthy et al. 1999; Petrenko et al. 1999; Rosen 2004). Additionally, CD93 is subject to metallo-protease-mediated ecto-domain cleavage or shedding, which is characteristic of several inflammatory mediators and adhesion molecules including TNF-α, TGF-α, TGF-β, EGF, CD44, and L-selectin (Bohlson et al. 2005). Despite its initial identification as a receptor for the C1q component of complement and demonstration of an in vivo kinetic defect in the clearance of apoptotic cells in B6 CD93−/− mice, the exact in vivo function of this molecule is yet to be elucidated.
Here, we identify a point mutation in the NOD Cd93 gene, which maps to the Idd13 locus, a region encoding a high degree of penetrance for diabetes susceptibility in non-obese diabetic (NOD) mice (Kim et al. 2000; Serreze et al. 1998). Interestingly, we also identified this Cd93 polymorphism in NZB/W F1 mice, to which the lupus susceptibility loci, Wbw1 and Nkt2, are tightly linked (Rahman et al. 2002; Esteban et al. 2003). This point mutation is associated with aberrant expression of CD93 on NOD and NZB/W F1 B cells in the pro-/pre-, immature, and TR subsets as compared with non-autoimmune B6 mice. The Idd13 and Wbw1 and Nkt2 loci are known to play a role in the regulation of the invariant natural killer T (iNKT) cell compartment (Chen et al. 2007; Esteban et al. 2003; Jordan et al. 2004, Rahman et al. 2002). Since NOD and NZB/W F1 mice are known to have deficiencies in their iNKT cell populations (Baxter et al. 1997; Cain et al. 2006; Chen et al. 2007; Duarte et al. 2004; Esteban et al. 2003; Jordan et al. 2004; Matsuki et al. 2003; Poulton et al. 2001; Rahman et al. 2002; Wagner et al. 2005), we sought to determine whether our finding of a mutation in the NOD and NZB/W F1 Cd93 gene is linked to regulation of iNKT cells in these autoimmune-prone strains. We assessed the iNKT cell populations in non-autoimmune B6 CD93 knockout (B6 CD93−/−) mice and found that they exhibit a profound state of CD4+ iNKT cell deficiency. Moreover, we found that the congenic B6.NODIdd13 mice, which carry the NOD Cd93 gene, also exhibited this CD4+ iNKT cell deficiency. These findings suggest that Cd93 may be an important autoimmune susceptibility gene influencing iNKT cell homeostasis.
Materials and methods
Mice
C57BL/6J, NOD/ShiLtJ, C3H/HeJ, BALB/cJ, NZBWF1/J, B6.NODIdd13-(D2Mit274-D2Mit343) (Jax#3064), NOD.B6Idd13-(D2Mit490-Ada) (Jax#2346), NOD.B6Idd13-(Zfp106-Il1a) (Jax#3050), and NOD.B6Idd13-(Il1a-Pcna) (Jax#3051) mice were obtained from Jackson Laboratories (Bar Harbor, ME). B6 CD93−/− mice, constructed from 129-derived ES cells, were generously donated by Dr. M. Botto and were backcrossed onto the C57BL/6 background for seven generations. All animals used in the described experiments were between 5 and 20 weeks old and were housed in specific pathogen-free conditions at the University of Pennsylvania Medical Center. Animal procedures were in accordance with the Animal Welfare Act.
Abs and flow cytometric analysis
The antibodies used in this study were as follows: PerCP conjugated anti-CD45R (B220, RA3-6B2), APC conjugated anti-IgM (II/41), FITC/PE conjugated anti-CD21/35 (7G6), PE conjugated anti-AA4.1, biotinylated 493 mAb (BD Biosciences, San Diego, CA), mAb1696 (R&D systems, Minneapolis, MN), and anti-Rat IgG2b (RG7/11.1) (BD Biosciences). Biotinylated mAb were detected by streptavidin-allophycocyanin (BD Biosciences). PE conjugated CD1d tetramer loaded with PBS-57 (Alpha-galactosyl ceramide (alpha-GalCer) analog) was prepared by the NIH Tetramer Facility. A total of 1–2 × 106 splenocytes, thymocytes, BM, or peripheral blood lymphocytes (PBL) derived cells from age- and sex-matched mice were surface stained in 96-well microtiter plates with various combinations of the previously described antibodies. Preparation and staining of livers was done as previously described (Hammond et al. 2001). Fluorescence-activated cell sorting (FACS) analysis was performed using a FACSCalibur (BD Biosciences, San Jose, CA), and the data were analyzed using FlowJo Software (version 8, Tree Star, Inc, Ashland, OR).
Bone-marrow-derived macrophages
Bone-marrow-derived macrophages were prepared from bone marrow progenitors. Briefly, bone marrow was flushed from femurs of 5- or 10-week-old mice and plated in cell culture dishes with Dulbecco’s modified Eagle’s medium (Invitrogen, San Diego, CA) supplemented with 15% L929 cell-conditioned medium (as a source of macrophage colony-stimulating factor), 10% heat-inactivated fetal calf serum, 100 U/ml penicillin/streptomycin (Invitrogen), and 10 mM HEPES. Additional media was added on day 4, and cells were incubated at 37°C in 5% CO2 for 7–8 days until uniform layers of macrophages were established.
sCD93 enzyme-linked immunosorbent assay
Blood was collected by cardiac puncture, placed on ice, centrifuged for 10 min at 3,200 rpm at 4°C, then serum was stored at −70°C until use. Mouse serum was assayed for soluble CD93 using CD93 sandwich enzyme-linked immunosorbent assay (ELISA). Briefly, Immulon 2 HB plates (Thermo Labsystems, Franklin, MA) were coated overnight at 4°C with 2 mcg/ml rat monoclonal anti-mouse CD93 (mAB1696, R&D Systems, Minneapolis, MN) in 0.1 M carbonate buffer, pH 7.4, washed with phosphate buffer saline containing 0.05% Tween-20 (PBST), and blocked with PBST containing 3% dried milk (block buffer) for 1 h. Diluted serum samples were added to wells and incubated for 1 h. After washing with PBST, 1 mcg/ml sheep anti-mouse CD93 antibody (AF1696, R&D Systems) was added to wells for 1 h followed by incubation with anti-sheep horseradish peroxidase-conjugated streptavidin (Jackson, Immunoresearch, West Grove, PA) diluted 1:2,000 in 1% dried milk. Sigma fast o-phenylenediamine dihydrochloride peroxidase substrate (Sigma-Aldrich, St. Louis, MO) was added, and the change in absorbance at 405 nm was measured.
Western blot
Bone-marrow-derived macrophages or B-cell precursors (purified with magnetic activated cell separation using B-cell isolation kit, Miltenyi Biotec, Auburn, CA) were harvested into lysis buffer (50 mM HEPES, pH 7.0, 150 mM NaCl, 10% glycerol, 1.2% Triton X-100, 1.5 mM MgCl2, 10 mM sodium pyrophosphate, 100 mM NaF, 1.25 mM sodium orthovanadate, and 1 mM phenylmethylsulfonyl fluoride plus 0.15 U/ml aprotinin, 10 mcg/ml leupeptin, 10 mcg/ml pepstatin A) and incubated on ice for 30 min. After incubation, lysates were centrifuged at 14,000×g for 10 min. The supernatants were collected, and protein concentrations were determined using the BCA kit (Pierce Biotechnology, Rockford, IL). Soluble cell extracts were then separated by 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF). The membrane was blocked overnight with 5% dry milk in TBST (0.05% Tween-20 in 20 mM Tris, pH 7.4, 150 mM NaCl). Lysates from bone-marrow-derived macrophages were probed for 2 h at room temperature with polyclonal anti-CD93 cytoplasmic tail Ab 1150 generated against C-terminal 11 aa of CD93 as previously described (Webster et al. 2000) and were washed and probed with secondary horseradish peroxidase conjugate for 1 h at room temperature and developed using ECL (Amersham Biosciences). Lysates from bone-marrow-derived B-cell precursors were incubated overnight with a polyclonal anti-CD93 (Anti-mouse C1qR1, R&D Systems) and probed with secondary anti-sheep antibody.
Polymerase chain reaction amplification and genomic sequencing
Genomic DNA was isolated from tail snip samples according to standard protocols. The CD93 gene was polymerase chain reaction (PCR) amplified from genomic DNA using primers designed from the published cDNA sequence (NCBI accession number NM_010740.3). The forward oligonucleotide used was 5′-ATGGCCATCTCAACTGGTTT-3′, and the reverse oligonucleotide used was 5′-TCAGCAGTCTGTCCCTGGTG-3′. The PCR product of 1,835 bp, covering the first EGF-like domain of CD93, was isolated on an agarose gel. The bands were cut, and purified DNA was extracted using the QIAGEN QIAquick Gel Extraction kit. Samples were sent for sequencing at the University of Pennsylvania School of Medicine DNA Sequencing Facility.
Statistical analysis
Statistical analysis was performed using the Mann–Whitney test, with differences considered significant at p < 0.05.
Results
The NOD and NZB/W F1 Cd93 alleles carry an Asn→His polymorphism at amino acid 264 in its first EGF-like domain
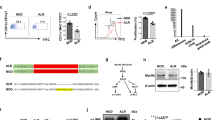
Genomic Cd93 DNA from B6, BALB/c, NOD, and NZB/W F1 mice was PCR amplified and sequenced using various primers to walk along the amplified DNA. A hitherto unidentified coding polymorphism at cDNA nucleotide position 790, which converts AAC→CAC, was discovered in NOD and NZB/W F1 mice (Fig. 1a). This mutation causes an amino acid substitution from Asn→His at position 264 in the first EGF-like domain of Cd93 (Fig. 1a).
Identification of a coding polymorphism in the first EGF-like domain of Cd93 in NOD and NZB/W F1 mice. a The Cd93 gene was PCR amplified from genomic DNA. Sequencing revealed a point mutation at cDNA nucleotide position 790. The AAC→CAC codon change in NOD and NZB/W F1 mice led to an amino acid substitution in the first EGF-like domain of Cd93 at position 264. The first EGF-like domain in which the defined polymorphism resides is stippled. b Bone marrow macrophage lysates (left) were separated on an 8% SDS-PAGE under reducing conditions, transferred to PVDF, and probed with 5 μg/ml polyclonal anti-CD93 cytoplasmic tail Ab 1150 (black arrow). Isolated bone marrow B-cell lysate immunoblots (right) were probed with 0.1 μg/ml polyclonal anti-CD93 (anti-C1qR1). The blots were stripped and re-probed for β-actin or β-tubulin to control for similar protein loading. The blots are from one experiment, representative of two, using different animals as a source of bone-marrow-derived macrophages or B cells. c sCD93 is detected in sera from NOD and NZB/W F1 mice. Sera were diluted (1:50) and tested for the presence of sCD93 by sandwich ELISA. Shown is the average of triplicate wells (± SD) from one experiment, representative of two, using different animals as a source of sera
CD93 protein is expressed in cell lysates and serum of NOD and NZB/W F1 mice
We next sought to determine whether CD93 protein is expressed in NOD and NZB/W F1 mice. Lysates from in vitro-differentiated, BM-derived macrophages were prepared and used in Western blot. CD93 protein was detected in both NOD and NZB/W F1 lysates as assessed using polyclonal anti-CD93 cytoplasmic tail Ab 1150 (Webster et al. 2000). CD93 bands were seen with NOD and NZB/W F1 mice comparable to the B6 and NOR control strains (Fig. 1b). Lysates of BM-derived macrophages from the B6 CD93−/− strain (Norsworthy et al. 2004) were used for a negative control. We performed a Western blot on purified BM-derived B-cell precursors to determine whether CD93 protein is detectable on B-cell precursors using a polyclonal anti-CD93 Ab. B-cell lysates from NOD and B6.NODIdd13 mice exhibited a CD93 band similar to B6 counterparts (Fig. 1b).
CD93 is subject to ecto-domain cleavage by metalloproteinase and, as such, exists in soluble form in vivo (Bohlson et al. 2005). Therefore, as another measure of protein expression, we assessed serum from NOD and NZB/W F1 mice for the presence of sCD93 in an ELISA assay. NOD and NZB/W F1 sera contained measurable levels of sCD93 at a slightly lower concentration compared to B6 sera (Fig. 1c). sCD93 levels did not change significantly with age. Additionally, the concentration of sCD93 in NOD and NZB/W F1 mice remained constant for up to 30 weeks of age, and no difference was noted between males and females (data not shown).
Surface CD93 expression by pro-/pre-, immature, and transitional B cells is deficient in NOD and NZB/W F1 mice
The expression of CD93 on the various B-cell developmental subsets was characterized using a previously described flow cytometric gating scheme (Cancro 2004; Quinn et al. 2006). In the BM, we resolved the pro-/pre-, immature, and mature recirculating B-cell subsets based on their expression of IgM and B220 (Fig. 2a). The B220lo/IgM− gate contains pro- and pre-B cells, the B220lo/IgM+ gate contains immature B cells, and the B220hi/IgM+ gate contains mature/recirculating B cells. In the peripheral blood and spleen, we resolved the transitional B-cell compartment using a B220+/IgM+/CD21/35lo gate and the mature/follicular subset using a B220+/IgM+/CD21/35int gate (Fig. 2a). CD93 is routinely used as an early B-cell developmental marker in non-autoimmune strains of mice (Cancro 2004). In B6 mice, CD93 is expressed at high levels on pro-/pre- and immature B-cell progenitors in the BM (Fig. 2a). Its expression continues on peripheral blood and splenic TR B cells; albeit at a lower level than that seen on B-cell progenitors in the BM (Fig. 2a). However, characterization of CD93 expression using the various B-cell developmental subsets in NOD mice revealed a deficiency in its expression by pro-/pre-, immature, and TR B cells in the BM, peripheral blood, and splenic compartments. Overall, cell surface staining of CD93 using the AA4.1 mAb revealed three- to sevenfold reduced expression on the surface of early NOD B-cell progenitors, as compared to their B6 counterparts (Fig. 2a). CD93 expression on early B-cell subsets of NZB/W F1 mice followed the staining pattern observed in NOD mice (data not shown). Both the absolute number and proportion of CD93+ B cells (i.e., AA4.1+) were significantly reduced in all early B-cell developmental subsets in NOD and NZB/W F1 mice, compared with B6 mice (Fig. 2b). We utilized three distinct mAbs to quantify the cell surface expression of CD93 in NOD and NZB/W F1 versus B6 mice (Fig. 2c). AA4.1, 493 and 1696 are known to bind CD93 (McKearn et al. 1984; Rolink et al. 1998). AA4.1, 493 and 1696 staining are deficient in B-cell progenitors of NOD mice as compared to their B6 counterparts (Fig. 2c). Our inability to detect surface CD93 in NOD and NZB/W F1 mice using anti-CD93 mAbs, despite the presence of CD93 protein expression in BM-derived B-cell precursors and macrophages using polyclonal anti-CD93 antibodies (Fig. 1b), is likely due to a conformation polymorphism in Cd93.
CD93 expression and absolute numbers on the various B-cell developmental subsets in BM and peripheral lymphoid organs. a The first column divides lymphoid-gated B cells in the bone marrow into mature recirculating (B220hi/IgM+), immature (B220lo/IgM+), and pro-/pre-B cells (B220lo/IgM−). The histograms resolve CD93 expression by AA4.1 antibody on pro-/pre- and immature B cells (black line) in contrast with mature recirculating B cells (dashed line). The ratio of the mean fluorescence indices (MFIs) of AA4.1 expression by pro-/pre- and immature B cells to the AA4.1 expression by mature recirculating B cells is shown as MFI fold Δ on the histograms. Splenic B-cell developmental subsets are divided into the marginal zone (MZ) pool (B220+ gated, IgMhi/CD21/35hi), FO pool (B220+ gated, IgMint/CD21/35int), and TR pool (B220+ gated, IgM+/CD21/35lo). The histograms display AA4.1 expression by the TR pool (black line) in comparison with the FO pool (dashed line). The ratio of MFIs of AA4.1 expression by TR B cells and FO B cells is shown as MFI fold Δ. B cells in the peripheral blood are divided into the mature recirculating (B220+ gated, IgM+/CD21/35int) and the TR pool (B220+ gated, IgM+/CD21/35lo). The histograms display AA4.1 expression by the TR pool (black line) in comparison with the mature recirculating pool (dashed line). The MFI fold Δ is an average of at least n = 5 mice per strain shown, and *p < 0.05 compared with B6 mice. b The left panel shows the total numbers of AA4.1+ pro-/pre- and immature B cells in the BM of B6 (n = 12; black bar) and NOD (n = 10; white bar) and NZB/W F1 (n = 5, gray bar) mice. The middle panel shows the total number AA4.1+ TR B cells in the spleen of B6 (n = 9; black bar), NOD (n = 9; white bar), and NZB/W F1 (n = 5; gray bar) mice. The right panel shows the percent AA4.1+ cells in the pro-/pre- and immature B cells in the BM and the TR B-cell compartments in the PBL and spleen of B6 (n = 12 for BM compartments, n = 9 for spleen, n = 5 for PBL), NOD (n = 10 for BM compartments, n = 9 for spleen, n = 5 for PBL), and NZB/W F1 (n = 5 for all compartments) mice. Error bars indicate ± SD, **p < 0.01 compared with B6 mice. c The column on the left shows characteristic AA4.1 expression of B220 cells in the BM of a B6, NOD, and NZB/W F1 mouse. On the right, AA4.1 and 493 Abs are used in conjunction on B220+ gated lymphocytes in the BM. The histograms show expression of mAb1696 by pro-/pre- and immature (black line) in contrast with mature recirculating (dashed line) B cells in the BM of B6, NOD, and NZB/W F1 mice. Staining is representative of at least n = 5 per mouse strain
The NOD CD93 defect maps to Idd13
The Cd93 gene maps to 84 cM on murine chromosome 2 (Kim et al. 2000) and falls within the NOD Idd13 locus (Serreze et al. 1998). Idd13 congenic B6 and NOD mice were obtained from Jackson Laboratories in order to establish whether these mice carry the B6 or NOD allele. Figure 3a summarizes the chromosomal regions introgressed from either the B6 or NOD strains onto the NOD and B6 backgrounds for each Idd13 congenic strain, respectively. The B6.NODIdd13 strain carries the NOD chromosome 2 from 52.5 to 84.2 cM. Early B-cell progenitors from the B6.NODIdd13 strain are deficient in CD93 expression as compared to B6 wild-type counterparts, similarly to NOD wild-type mice (Fig. 3b). Three NOD.B6Idd13 subcongenic mouse lineages (JAX# 2346, 3050, and 3051) are currently available from Jackson Laboratories (Fig. 3a). The NOD.B6Idd13-(D2Mit490-Ada) strain harbors B6 chromosome 2 in the range of 64.5–94 cM and, as such, carries the B6 Cd93 allele (Serreze et al. 1998). Early B-cell progenitors in this strain express CD93 similarly to B6 wild-type counterparts (Fig. 3b). The NOD.B6Idd13-(Zfp106-Il1a) and NOD.B6Idd13-(Il1a-Pcna) strains harbor B6 chromosome 2 in the range of 67.2–73 and 73–75 cM, respectively (Serreze et al. 1998). Early B-cell progenitors from these Idd13 subcongenic strains are deficient in CD93 expression as compared to B6 wild-type counterparts, similarly to NOD wild-type mice (Fig. 3b).
The NOD CD93 defect maps to Idd13. a Chromosomal map showing regions of either NOD chromosome 2 (white bar) or B6 chromosome 2 (black bar) introgressed onto the B6 or NOD background for each congenic strain obtained from Jackson Laboratories. b The left column shows AA4.1 staining on B-cell progenitors (B220lo) in the BM for each congenic strain. The middle column divides B cells in the bone marrow into mature recirculating ( B220hi/IgM+), immature (B220lo/IgM+), and pro-/pre-B cells (B220lo/IgM−) for each of the congenic strains. The histograms resolve AA4.1 expression by pro-/pre- and immature B cells (black line) in contrast with mature recirculating B cells (gray line). The ratio of the mean fluorescence indices (MFIs) of AA4.1 expression by pro-/pre- and immature B cells to the AA4.1 expression by mature recirculating B cells is shown as MFI fold Δ. The MFI fold Δ is an average of at least n = 4 mice per strain shown, and *p < 0.05 compared with B6 mice
B6 CD93−/− mice and B6.NODIdd13 mice exhibit a profound state of CD4+ iNKT cell deficiency
The Cd93 locus maps to a region of chromosome 2, which regulates iNKT cell number and/or function (Chen et al. 2007; Esteban et al. 2003). Therefore, we undertook an analysis of the iNKT cell compartments of B6 CD93−/− (Norsworthy et al. 2004), B6.NODIdd13, and NOD mice. iNKT cells were analyzed in the liver, thymus and spleen using an alpha-GalCer analog, PBS57-loaded CD1d tetramer. iNKT cells were further resolved into CD4+ and CD4− subsets in the spleen and liver and CD4 single positive (CD4SP) and double-negative (DN) subsets in the thymus. Flow cytometric analysis of lymphoid cells in the liver of non-autoimmune B6 mice revealed 15.9 ± 5.79% of the population staining for iNKT cells (Fig. 4a). In contrast, there was a global deficiency of iNKT cells in lymphoid populations in the livers of B6 CD93−/−, B6.NODIdd13, and NOD mice (9.7 ± 4.20%, 7.71 ± 4.29%, and 8.15 ± 3.11%, respectively, p < 0.05 compared with B6 mice, Fig. 4a). We further resolved the population of iNKT cells into the CD4+ iNKT and CD4− iNKT cell subsets (Fig. 4a, right column), which allowed us to determine the frequencies of CD4+ iNKT and CD4− iNKT cells in the liver (Fig. 4b). There is a twofold reduction in the frequency of CD4+ hepatic iNKT cells in B6 CD93−/−, B6.NODIdd13, and NOD, compared with B6 mice (p < 0.05, Fig. 4b). Conversely, the frequency of CD4− iNKT cells in the liver is similar across each of the tested strains of mice (p > 0.05). Figure 4c shows that the total number of thymic DN and CD4SP iNKT cells is up to twofold less in B6 CD93−/−, B6.NODIdd13, and NOD mice, compared with B6 (p < 0.05). Similarly, there is a two- to threefold reduction of total splenic CD4+ iNKT cells in B6 CD93−/−, B6.NODIdd13, and NOD compared with B6 mice (p < 0.05), while the CD4− iNKT cell numbers is comparable in all strains of mice tested (p > 0.05, Fig. 4d). To exclude the possibility that the reduction of CD4+ iNKT cells was due to an overall lymphopenia, we analyzed the total numbers of thymic and splenic lymphocytes and found that the total number of lymphoid cells in B6, B6 CD93−/−, B6.NODIdd13, and NOD mice was comparable (p > 0.05, data not shown). Our studies thus confirm that NOD mice possess a CD4+ iNKT cell deficiency (Baxter et al. 1997; Godfrey et al. 1997; Hammond et al. 2001) and show that B6 CD93−/− and B6.NODIdd13 mice exhibit a CD4+ iNKT cell deficiency similar to NOD mice.
iNKT cell analysis in B6, B6 CD93−/−, B6.NODIdd13 and NOD mice. a The left panel shows the frequency of iNKT cell staining (CD3lo, PBS57-loaded CD1d Tetramer+) in the liver of B6 (n = 9, mean 15.9 ± 5.79%) B6 CD93−/− (n = 7, mean 9.78 ± 4.20%), B6.NODIdd13, (n = 5, mean 7.71 ± 4.29%), and NOD (n = 5, mean 8.15 ± 3.11%) mice. The right panel further resolves CD4+ iNKT and CD4− iNKT cells. b The bar graph indicates the frequency of CD4+ and CD4− iNKT (CD3lo, PBS57-loaded CD1d Tetramer+) cells in the livers of B6 CD93−/−, B6.NODIdd13, and NOD mice. c The bar graph shows the absolute number of thymic DN and CD4SP iNKT cells (CD3lo, PBS57-loaded CD1d Tetramer+) in B6, B6 CD93−/−, B6.NODIdd13, and NOD mice. d The bar graph indicates the total number of splenic CD4+ and CD4− iNKT (CD3lo, PBS57-loaded CD1d Tetramer+) cells in B6, B6 CD93−/−, B6.NODIdd13, and NOD mice. Error bars indicate ± SD, *p < 0.05 compared with B6 mice
Discussion
In the present study, we identify a hitherto unknown Cd93 polymorphism in NOD mice. The Cd93 locus is tightly linked to the diabetes susceptibility gene, Idd13, and the Lupus susceptibility loci, Wbw1 and Nkt2, in the region of 82–84 cM on mouse chromosome 2 (Esteban et al. 2003; Jordan et al. 2004; Rahman et al. 2002). Both Idd13 and Nkt2 loci have been implicated in regulating the differentiation of natural killer T (NKT) cells, whose relative deficiency in NOD and NZB/W F1 mice is thought to play an important role in the pathogenesis of islet inflammation (Cain et al. 2006; Chen et al. 2007; Duarte et al. 2004; Esteban et al. 2003; Jordan et al. 2004; Matsuki et al. 2003; Poulton et al. 2001; Rahman et al. 2002; Wagner et al. 2005). A subcongenic analysis by Chen et al. revealed that there are at least two genes within Idd13 that regulate iNKT cell numbers and that NOD mice congenic for the B6 Idd13 locus were found to be protected from autoimmune diabetes progression associated with normalization of the NOD iNKT cell deficient phenotype (Chen et al. 2007). Our data show that B6.NODIdd13 congenic mice, which carry the NOD Cd93 allele, are characterized by a deficit in the CD4+ iNKT cell compartment. However, given the large span of chromosomal DNA introgressed into these congenic mice, Cd93 is only one among many potential candidate genes driving the iNKT cell phenotype. Fletcher et al. have proposed a list of 19 candidate genes, including Cd93, within the Idd13 region, which may be responsible for correcting the NOD NKT cell deficiency. Through construction of NOD mice with a congenic insertion of 12.6 Mb of B6-derived sequence within Idd13, the authors show partial correction of the type 1 NKT cell defect in NOD mice (Fletcher et al. 2008). Notably, Fletcher et al. also confirm the genetic location of the Nkt2 NKT cell control gene to chromosome 2 and provide evidence of the involvement of more than one locus in this region. Given the presence of an NOD and NZB/W F1 specific polymorphism in Cd93, we questioned whether this gene might be involved in the regulation of iNKT cell homeostasis. We therefore obtained B6 CD93−/− mice and analyzed their iNKT cell compartments. These analyses revealed a profound deficiency in the CD4+ subset of iNKT cells in CD93−/− mice. It should be noted that the B6 CD93−/− mice were constructed using 129 ES cells. Thus, it remains a possibility that 129 derived alleles within the Idd13 locus, other than Cd93, are identical to their NOD counterparts and cause the observed reduction in CD4+ iNKT cells. In the vicinity of Cd93 (1 Mb upstream and downstream), 43% of single-nucleotide polymorphisms (SNPs) in NOD and 129 mice are homologous. However, among the homologous SNPs, 84% are also homologous with B6 alleles and can thus be assumed to not contribute to iNKT cell defects (SNP analysis performed via Mouse Genome Informatics; http://www.informatics.jax.org). Furthermore, at backcross 7, our CD93−/− mice are 99.22% genetically identical with B6 mice. While we believe it unlikely that there are a significant proportion of remaining 129 derived loci contributing to the observed CD4+ iNKT cell deficit in CD93−/− mice, we acknowledge it as an important caveat.
Several previous reports had noted an overall deficiency in AA4.1 (i.e., CD93) expression in the bone marrow of NOD mice (Langmuir et al. 1993; Quinn et al. 2006; Silveira and Grey 2006). However, these studies did not further elucidate the specific cell populations with the deficit or its basis and functional significance. Here, using three distinct mAbs specific for mouse CD93, we demonstrated a marked defect in the staining pattern of CD93, most prominently visible on early B-cell developmental subsets in the bone marrow. Furthermore, we demonstrated that NZB/W F1 mice exhibited a similar defect in CD93 expression on early B-cell subsets. Despite this aberrant staining pattern observed via flow cytometry, we detected CD93 protein expression using polyclonal antibodies in Western blot assays of BM-derived macrophages or purified B-cell precursors in NOD or NZB/W F1 mice. Moreover, soluble CD93 was detectable in the serum of NOD and NZB/W F1 mice at a level comparable to that seen in non-autoimmune mice. Importantly, deficient CD93 staining using three distinct mAbs was associated with an Asn→His polymorphism at amino acid 264 in NOD and NZB/W F1 mice. This polymorphism falls within the first EGF-like domain of Cd93. We believe it likely that this mutation causes a conformational change that renders NOD and NZB/W F1 CD93 virtually undetectable by these monoclonal antibodies.
CD93 was originally isolated and cloned during a search for myeloid surface molecules involved in the enhancement of C1q mediated phagocytosis (Nepomuceno et al. 1997). However, macrophages from CD93-deficient mice are not deficient in their response to C1q. These mice do, however, exhibit an impaired uptake of apoptotic cells in vivo indicating that this receptor contributes to the removal of dying cells (Rolink et al. 1998). In this context, it has been suggested that the priming of autoreactive T and B cells in both autoimmune diabetes and lupus involves an initial response to apoptotic target cells (Turley et al. 2003). Thus, it may be important to determine whether the Cd93 polymorphism in NOD and NZB/W F1 mice leads to inefficient clearance of apoptotic bodies, thereby setting the stage for the initiation of an autoimmune response. In examining the candidacy of Cd93 as an Idd13 candidate gene, it will be important to also consider its recognized role in promoting the uptake of apoptotic cells and immune complexes by antigen presenting cells.
Our recognition of the NOD Cd93 mutation was a fortuitous outcome of our phenotypic characterization of NOD B-cell development (Quinn et al. 2006). It is well established that CD93 is a marker for early B-cell developmental subsets (Cancro 2004). The relationship between CD93 expression by early B-cell progenitors and iNKT cell homeostasis remains an active area of investigation in our laboratory. Nevertheless, it is possible that expression of this protein on developing B cells plays an independent role in the regulation of B-cell compartment homeostasis. Indeed, it has been shown that mutations within the EGF-like domains of the selectin family members, to which CD93 bears a unique resemblance, inhibit their activity as adhesion molecules (Kansas 1996; Rosen 2004). It will therefore be important to determine whether the NOD polymorphism imparts a defect in immature/TR B-cell migration dynamics as we previously demonstrated (Quinn et al. 2006).
Recently, CD93 expression was shown to be re-induced during plasma cell differentiation from primary B cells (Chevrier et al. 2009). Furthermore, Chevrier et al. suggest that while CD93-deficient mice make normal humoral responses following immunization, they are unable to maintain long-lived plasma cells (LLPC) within the bone-marrow niche (Chevrier et al. 2009). Whether this phenotype is related to the iNKT cell deficiency seen in these mice remains to be determined. Nonetheless, it has been shown that mice lacking iNKT cells cannot sustain antibody responses and LLPC populations (Galli et al. 2007). Therefore, it will be important to determine whether the reliance of LLPC on CD93 is related to their iNKT cell deficient phenotype. Additionally, the CD1 restricted NKT cell specific mitogen alpha-GalCer can induce antibody responses as well as B-cell memory (Galli et al. 2007). iNKT help to B cells can occur without cognate interaction, provided that B cells express the costimulatory molecule, CD40 (Tonti et al. 2009). We are currently investigating the interplay between CD93, iNKT cells, and B cells in both early development and later as they differentiate into LLPCs.
In summary, we describe a coding polymorphism in the first EGF-like domain of Cd93. This gene maps to a diabetes susceptibility locus on chromosome 2 and is also tightly linked to the NZB/W F1 Wbw1 and Nkt2 disease susceptibility loci, which are thought to regulate NKT cells. Indeed, CD93 deficiency was found to promote a profound state of CD4+ iNKT cell lymphopenia. These data suggest that Cd93 may be an autoimmune susceptibility gene and may play an important role in regulating the CD4+ iNKT cell compartment. These findings will direct future studies to determine whether Cd93 is a disease susceptibility gene in NOD and NZB/W F1 mice and whether similar polymorphisms exist in type 1 diabetes or lupus patients.
Abbreviations
- BM:
-
Bone marrow
- TR:
-
Transitional
- MZ:
-
Marginal zone
- iNKT cell:
-
Invariant natural killer T cell
- NOD:
-
Non-obese diabetic
- DN:
-
Double negative
- CD4SP:
-
CD4 single positive
- Idd:
-
Insulin-dependent diabetes
- alpha-GalCer:
-
Alpha-galactosyl ceramide
- LLPC:
-
Long-lived plasma cell
References
Baxter AG, Kinder SJ, Hammond KJ, Scollay R, Godfrey DI (1997) Association between alphabetaTCR+CD4-CD8-T-cell deficiency and IDDM in NOD/Lt mice. Diabetes 46(4):572–582
Bohlson SS, Silva R, Fonseca MI, Tenner AJ (2005) CD93 is rapidly shed from the surface of human myeloid cells and the soluble form is detected in human plasma. J Immunol 175(2):1239–1247
Cain JA, Smith JA, Ondr JK, Wang B, Katz JD (2006) NKT cells and IFN-gamma establish the regulatory environment for the control of diabetogenic T cells in the nonobese diabetic mouse. J Immunol 176(3):1645–1654
Cancro MP (2004) Peripheral B-cell maturation: the intersection of selection and homeostasis. Immunol Rev 197:89–101
Chen YG, Driver JP, Silveira PA, Serreze DV (2007) Subcongenic analysis of genetic basis for impaired development of invariant NKT cells in NOD mice. Immunogenetics 59(9):705–712
Chevrier S, Genton C, Kallies A, Karnowski A, Otten LA, Malissen B, Malissen M, Botto M, Corcoran LM, Nutt SL, Acha-Orbea H (2009) CD93 is required for maintenance of antibody secretion and persistence of plasma cells in the bone marrow niche. Proc Natl Acad Sci U S A 106:3895–3900
Danet GH, Luongo JL, Butler G, Lu MM, Tenner AJ, Simon MC, Bonnet DA (2002) C1qRp defines a new human stem cell population with hematopoietic and hepatic potential. Proc Natl Acad Sci U S A 99(16):10441–10445
Dean YD, McGreal EP, Gasque P (2001) Endothelial cells, megakaryoblasts, platelets and alveolar epithelial cells express abundant levels of the mouse AA4 antigen, a C-type lectin-like receptor involved in homing activities and innate immune host defense. Eur J Immunol 31(5):1370–1381
Duarte N, Stenstrom M, Campino S, Bergman ML, Lundholm M, Holmberg D, Cardell SL (2004) Prevention of diabetes in nonobese diabetic mice mediated by CD1d-restricted nonclassical NKT cells. J Immunol 173(5):3112–3118
Esteban LM, Tsoutsman T, Jordan MA, Roach D, Poulton LD, Brooks A, Naidenko OV, Sidobre S, Godfrey DI, Baxter AG (2003) Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol 171(6):2873–2878
Fletcher JM, Jordan MA, Snelgrove SL, Slattery RM, Dufour FD, Kyparissoudis K, Besra GS, Godfrey DI, Baxter AG (2008) Congenic analysis of the NKT cell control gene Nkt2 implicates the peroxisomal protein Pxmp4. J Immunol 181(5):3400–3412
Fonseca MI, Carpenter PM, Park M, Palmarini G, Nelson EL, Tenner AJ (2001) C1qR(P), a myeloid cell receptor in blood, is predominantly expressed on endothelial cells in human tissue. J Leukoc Biol 70:793–800
Galli G, Pittoni P, Tonti E, Malzone C, Uematsu Y, Tortoli M, Maione D, Volpini G, Finco O, Nuti S, Tavarini S, Dellabona P, Rappuoli R, Casorati G, Abrignani S (2007) Invariant NKT cells sustain specific B cell responses and memory. Proc Natl Acad Sci U S A 104(10):3984–3989
Godfrey DI, Kinder SJ, Silvera P, Baxter AG (1997) Flow cytometric study of T cell development in NOD mice reveals a deficiency in alphabetaTCR+CDR-CD8- thymocytes. J Autoimmun 10(3):279–285
Hammond KJ, Pellicci DG, Poulton LD, Naidenko OV, Scalzo AA, Baxter AG, Godfrey DI (2001) CD1d-restricted NKT cells: an interstrain comparison. J Immunol 167(3):1164–1173
Jordan MA, Fletcher J, Baxter AG (2004) Genetic control of NKT cell numbers. Immunol Cell Biol 82(3):276–284
Kansas GS (1996) Selectins and their ligands: current concepts and controversies. Blood 88(9):3259–3287
Kim TS, Park M, Nepomuceno RR, Palmarini G, Winokur S, Cotman CA, Bengtsson U, Tenner AJ (2000) Characterization of the murine homolog of C1qR(P): identical cellular expression pattern, chromosomal location and functional activity of the human and murine C1qR(P). Mol Immunol 37(7):377–389
Langmuir PB, Bridgett MM, Bothwell AL, Crispe IN (1993) Bone marrow abnormalities in the non-obese diabetic mouse. Int Immunol 5(2):169–177
Matsuki N, Stanic AK, Embers ME, Van Kaer L, Morel L, Joyce S (2003) Genetic dissection of V alpha 14J alpha 18 natural T cell number and function in autoimmune-prone mice. J Immunol 170(11):5429–5437
McKearn JP, Baum C, Davie JM (1984) Cell surface antigens expressed by subsets of pre-B cells and B cells. J Immunol 132(1):332–339
Nepomuceno RR, Tenner AJ (1998) C1qRP, the C1q receptor that enhances phagocytosis, is detected specifically in human cells of myeloid lineage, endothelial cells, and platelets. J Immunol 160(4):1929–1935
Nepomuceno RR, Henschen-Edman AH, Burgess WH, Tenner AJ (1997) cDNA cloning and primary structure analysis of C1qR(P), the human C1q/MBL/SPA receptor that mediates enhanced phagocytosis in vitro. Immunity 6(2):119–129
Norsworthy PJ, Taylor PR, Walport MJ, Botto M (1999) Cloning of the mouse homolog of the 126-kDa human C1q/MBL/SP-A receptor, C1qR(p). Mamm Genome 10(8):789–793
Norsworthy PJ, Fossati-Jimack L, Cortes-Hernandez J, Taylor PR, Bygrave AE, Thompson RD, Nourshargh S, Walport MJ, Botto M (2004) Murine CD93 (C1qRp) contributes to the removal of apoptotic cells in vivo but is not required for C1q-mediated enhancement of phagocytosis. J Immunol 172(6):3406–3414
Petrenko O, Beavis A, Klaine M, Kittappa R, Godin I, Lemischka IR (1999) The molecular characterization of the fetal stem cell marker AA4. Immunity 10(6):691–700
Poulton LD, Smyth MJ, Hawke CG, Silveira P, Shepherd D, Naidenko OV, Godfrey DI, Baxter AG (2001) Cytometric and functional analyses of NK and NKT cell deficiencies in NOD mice. Int Immunol 13(7):887–896
Quinn WJ 3rd, Noorchashm N, Crowley JE, Reed AJ, Noorchashm H, Naji A, Cancro MP (2006) Cutting edge: impaired transitional B cell production and selection in the nonobese diabetic mouse. J Immunol 176(12):7159–7164
Rahman ZS, Tin SK, Buenaventura PN, Ho CH, Yap EP, Yong RY, Koh DR (2002) A novel susceptibility locus on chromosome 2 in the (New Zealand Black x New Zealand White)F1 hybrid mouse model of systemic lupus erythematosus. J Immunol 168(6):3042–3049
Rolink AG, Andersson J, Melchers F (1998) Characterization of immature B cells by a novel monoclonal antibody, by turnover and by mitogen reactivity. Eur J Immunol 28(11):3738–3748
Rosen SD (2004) Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol 22:129–156
Serreze DV, Bridgett M, Chapman HD, Chen E, Richard SD, Leiter EH (1998) Subcongenic analysis of the Idd13 locus in NOD/Lt mice: evidence for several susceptibility genes including a possible diabetogenic role for beta 2-microglobulin. J Immunol 160(3):1472–1478
Silveira PA, Grey ST (2006) B cells in the spotlight: innocent bystanders or major players in the pathogenesis of type 1 diabetes. Trends Endocrinol Metab 17(4):128–135
Tonti E, Galli G, Malzone C, Abrignani S, Casorati G, Dellabona P (2009) NKT-cell help to B lymphocytes can occur independently of cognate interaction. Blood 113(2):370–376
Turley S, Poirot L, Hattori M, Benoist C, Mathis D (2003) Physiological beta cell death triggers priming of self-reactive T cells by dendritic cells in a type-1 diabetes model. J Exp Med 198(10):1527–1537
Wagner MJ, Hussain S, Mehan M, Verdi JM, Delovitch TL (2005) A defect in lineage fate decision during fetal thymic invariant NKT cell development may regulate susceptibility to type 1 diabetes. J Immunol 174(11):6764–6771
Webster SD, Park M, Fonseca MI, Tenner AJ (2000) Structural and functional evidence for microglial expression of C1qR(P), the C1q receptor that enhances phagocytosis. J Leukoc Biol 67(1):109–116
Zhang M, Bohlson SS, Dy M, Tenner AJ (2005) Modulated interaction of the ERM protein, moesin, with CD93. Immunology 115:63–73
Acknowledgements
This study was supported by National Institutes of Health Grants K08-DK064603, R03-DK080286 (to H.N.) and AI-41090 (to A.T.) and Juvenile Diabetes Research Foundation Grant 4-2008-351 (A.N.). The authors thank Christopher Prieto, Aurada Cholapranee, Kumar Vivek, Laurie Lenox, Robert E. Roses, Ronald F. Parsons, Robert Redfield III, Ming Yu, Brigitte Koeberlein, and Margaret Pain for their technical and scientific input.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ghazal Zekavat and Raha Mozaffari contributed equally to this study.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Zekavat, G., Mozaffari, R., Arias, V.J. et al. A novel CD93 polymorphism in non-obese diabetic (NOD) and NZB/W F1 mice is linked to a CD4+ iNKT cell deficient state. Immunogenetics 62, 397–407 (2010). https://doi.org/10.1007/s00251-010-0442-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-010-0442-3