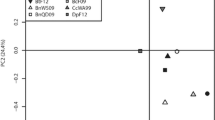
Abstract
The occurrence of interaction between insects and fungi is interesting from an ecological point of view, particularly when these interactions involve insect pests and plant pathogens within an agroecosystem. In this study, we aimed to perform an accurate analysis on the fungal microbiota associated to Bactrocera oleae (Rossi) through a metabarcoding approach based on 454 pyrosequencing. From this analysis, we retrieved 43,549 reads that clustered into 128 operational taxonomic units (OTUs), of which 29 resulted in the “core” associate fungi of B. oleae. This fungal community was mainly represented by sooty mould fungi, such as Cladosporium spp., Alternaria spp. and Aureobasidium spp., by plant pathogens like Colletotrichum spp. and Pseudocercospora spp., along with several other less abundant taxa whose ecology is unclear in most of the cases. Our findings lead to new insights into the microbial ecology of this specific ecological niche, enabling the understanding of a complex network of interactions within the olive agroecosystem.


Similar content being viewed by others
References
Boucias DG, Lietze VU, Teal P (2012) Chemical signals that mediate insect-fungal interactions. In: Biocommunication of Fungi. Springer, Dordrecht, pp 305–336
Six DL (2012) Ecological and evolutionary determinants of bark beetle—fungus symbioses. Insects 3:339–366
Hatcher PE (1995) Three‐way interactions between plant pathogenic fungi, herbivorous insects and their host plants. Biol Rev 70:639–694
Tack AJ, Dicke M (2013) Plant pathogens structure arthropod communities across multiple spatial and temporal scales. Funct Ecol 27:633–645
Ploetz RC, Hulcr J, Wingfield MJ, De Beer ZW (2013) Destructive tree diseases associated with ambrosia and bark beetles: black swan events in tree pathology? Plant Dis 97:856–872
Yousef M, Quesada-Moraga E, Garrido-Jurado I (2015) Compatibility of herbicides used in olive orchards with a Metarhizium brunneum strain used for the control of preimaginal stages of tephritids in the soil. J Pest Sci 88:605–612. doi:10.1007/s10340-014-0632-0
Shah P, Pell J (2003) Entomopathogenic fungi as biological control agents. Appl Microbiol Biotechnol 61:413–423
Malheiro R, Casal S, Baptista P, Pereira JA (2015) A review of Bactrocera oleae (Rossi) impact in olive products: from the tree to the table. Trends Food Sci Technol 44:226–242
Daane KM, Johnson MW (2010) Olive fruit fly: managing an ancient pest in modern times. Annu Rev Entomol 55:151–169
Bengochea P, Budia F, Viñuela E, Medina P (2014) Are kaolin and copper treatments safe to the olive fruit fly parasitoid Psyttalia concolor? J Pest Sci 87:351–359
Benelli G, Carpita A, Simoncini S, Raspi A, Canale A (2014) For sex and more: attraction of the tephritid parasitoid Psyttalia concolor (Hymenoptera: Braconidae) to male sex pheromone of the olive fruit fly, Bactrocera oleae. J Pest Sci 87:449–457
Benelli G, Daane KM, Canale A, Niu CY, Messing RH, Vargas RI (2014) Sexual communication and related behaviours in Tephritidae: current knowledge and potential applications for Integrated Pest Management. J Pest Sci 87:385–405
Campolo O, Palmeri V, Malacrinò A, Laudani F, Castracani C, Mori A, Grasso DA (2015) Interaction between ants and the Mediterranean fruit fly: new insights for biological control. Biol Control 90:120–127
Hsu JC, Huang LH, Feng HT, Su WY (2015) Do organophosphate-based traps reduce control efficiency of resistant tephritid flies? J Pest Sci 88:181–190
Schena L, Agosteo G, Cacciola SO (2011) Olive diseases and disorders. Transworld Research Network, Kerala
Malacrinò A, Schena L, Campolo O, Laudani F, Palmeri V (2015) Molecular analysis of the fungal microbiome associated with the olive fruit fly Bactrocera oleae. Fungal Ecol 18:67–74
Frisullo S, Carlucci A (2011) Minor fungal diseases of olives. In: Schena L, Agosteo G, Cacciola SO (eds) Olive diseases and disorders. Transworld Research Network, Kerala, pp 291–304
Cacciola S, Faedda R, Sinatra F, Agosteo G, Schena L, Frisullo S, di San Lio GM (2012) Olive anthracnose. J Plant Pathology 94:29–44
Nigro F, Ferrara M (2011) Olive Cercosporiosis. In: Schena L, Agosteo G, Cacciola SO (eds) Olive diseases and disorder. Transworld Research Network, Kerala, pp 247–258
Müller T, Ruppel S (2014) Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol Ecol 87:2–17
Taylor MW, Tsai P, Anfang N, Ross HA, Goddard MR (2014) Pyrosequencing reveals regional differences in fruit‐associated fungal communities. Environ Microbiol 16:2848–2858
Prigigallo MI, Abdelfattah A, Cacciola SO, Faedda R, Sanzani SM, Cooke DE, Schena L (2016) Metabarcoding analysis of Phytophthora diversity using genus-specific primers and 454 pyrosequencing. Phytopathology 106:305–313
Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E (2005) UNITE: a database providing web‐based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H (2010) ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:1
Ihrmark K, Bödeker IT, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandström-Durling M, Clemmensen KE (2012) New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 82:666–677
Prigigallo M, Mosca S, Cacciola S, Cooke D, Schena L (2015) Molecular analysis of Phytophthora diversity in nursery‐grown ornamental and fruit plants. Plant Pathol 64:1308–1319
Abdelfattah A, Nicosia MGLD, Cacciola SO, Droby S, Schena L (2015) Metabarcoding analysis of fungal diversity in the phyllosphere and carposphere of olive (Olea europaea). PLoS One 10, e0131069
Schena L, Cooke DE (2006) Assessing the potential of regions of the nuclear and mitochondrial genome to develop a “molecular tool box” for the detection and characterization of Phytophthora species. J Microbiol Methods 67:70–85
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols 18:315–322
Schmidt PA, Bálint M, Greshake B, Bandow C, Römbke J, Schmitt I (2013) Illumina metabarcoding of a soil fungal community. Soil Biol Biochem 65:128–132
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Reeder J, Knight R (2010) Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat Methods 7:668
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Bengtsson‐Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, Wit P, Sánchez‐García M, Ebersberger I, Sousa F (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919
Magurran AE, Henderson PA (2003) Explaining the excess of rare species in natural species abundance distributions. Nature 422:714–716
Knief C (2014) Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front Plant Sci 5:216
Abdelfattah A, Wisniewski M, Li Destri Nicosia MG, Cacciola SO, Schena L (2016) Metagenomic analysis of fungal diversity on strawberry plants and the effect of management practices on the fungal community structure of aerial organs. PLoS ONE 11(8):e0160470. doi:10.1371/journal.pone.0160470
Abdelfattah A, Wisniewski M, Droby S, Schena L (2016) Spatial and compositional variation in the fungal communities of organic and conventionally grown apple fruit at the consumer point-of-purchase. Horticulture Research 3:16042. doi:10.1038/hortres.2016.47
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Flessa F, Peršoh D, Rambold G (2012) Annuality of Central European deciduous tree leaves delimits community development of epifoliar pigmented fungi. Fungal Ecol 5:554–561
El-Morsy E, Dohlob S, Hyde K (2006) Diversity of Alternaria alternata a common destructive pathogen of Eichhornia crassipes in Egypt and its potential use in biological control. Fungal Divers 23:139–158
Woudenberg J, Groenewald J, Binder M, Crous P (2013) Alternaria redefined. Stud Mycol 75:171–212
Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M (2014) The sooty moulds. Fungal Divers 66:1–36
Damm U, Cannon P, Woudenberg J, Crous P (2012) The Colletotrichum acutatum species complex. Stud Mycol 73:37–113
Mosca S, Nicosia MGLD, Cacciola SO, Schena L (2014) Molecular analysis of Colletotrichum species in the carposphere and phyllosphere of olive. PLoS One 9, e114031
Faedda R, Agosteo GE, Schena L, Mosca S, Frisullo S, di San Lio GM, Cacciola SO (2011) Colletotrichum clavatum sp. nov. identified as the causal agent of olive anthracnose in Italy. Phytopathol Mediterr 50:283–302
Schena L, Mosca S, Cacciola S, Faedda R, Sanzani S, Agosteo G, Sergeeva V, Magnano di San Lio G (2014) Species of the Colletotrichum gloeosporioides and C. boninense complexes associated with olive anthracnose. Plant Pathol 63:437–446
Acknowledgments
This work was partially funded by SAF@MED (PON a3_00016)-PON Ricerca e competitività 2007-2013 and PON PON03PE_00090_2 “Modelli sostenibili e nuove tecnologie per la valorizzazione delle olive e dell’olio extravergine di oliva prodotto in Calabria”-PON Ricerca e competitività 2007–2013.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Authors’ Contribution
All authors designed the research. LS and VP supervised and coordinated the work. AM, FL, SM and OC provided samples. AM performed lab work, data analysis and data interpretation. AM wrote the first draft of the manuscript. All authors contributed to, corrected and approved the final manuscript.
Data Accessibility
Raw data from 454 sequencing are available on NCBI SRA database under the Project Accession ID: SRP089714. Sequences used for phylogenetic analysis are available as Supplementary material 2.
Rights and permissions
About this article
Cite this article
Malacrinò, A., Schena, L., Campolo, O. et al. A Metabarcoding Survey on the Fungal Microbiota Associated to the Olive Fruit Fly. Microb Ecol 73, 677–684 (2017). https://doi.org/10.1007/s00248-016-0864-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0864-z