Abstract
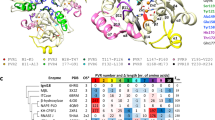
The serine β-lactamases present a special problem for phylogenetics because they have diverged so much that they fall into three classes that share no detectable sequence homology among themselves. Here we offer a solution to the problem in the form of two phylogenies that are based on a protein structure alignment. In the first, structural alignments were used as a guide for aligning amino acid sequences and in the second, the average root mean square distances between the alpha carbons of the proteins were used to create a pairwise distance matrix from which a neighbor-joining phylogeny was created. From those phylogenies, we show that the Class A and Class D β-lactamases are sister taxa and that the divergence of the Class C β-lactamases predated the divergence of the Class A and Class D β-lactamases.


Similar content being viewed by others
References
SF Altschul W Gish W Miller EW Myers DJ Lipman (1990) ArticleTitleBasic local alignment search tool. J Mol Biol 215 403–410 Occurrence Handle10.1006/jmbi.1990.9999 Occurrence Handle1:CAS:528:DyaK3MXitVGmsA%3D%3D Occurrence Handle2231712
SF Altschul TL Madden AA Schäffer J Zhang Z Zhang W Miller DJ Lipman (1997) ArticleTitleGapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 Occurrence Handle9254694
RP Ambler (1980) ArticleTitleThe structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 289 321–331
M Barlow BG Hall (2002a) ArticleTitleOrigin and evolution of the AmpC β-lactamases of Citrobacter freundii. Antimicrob Agents Chemother 46 1190–1198
M Barlow BG Hall (2002b) ArticleTitlePhylogenetic analysis shows that the OXA β-lactamase genes have been on plasmids for millions of years. J Mol Evol 55 314–321
R Breitling D Laubner J Adamski (2001) ArticleTitleStructure-based phylogenetic analysis of short-chain alcohol dehydrogenases and reclassification of the 17beta-hydroxysteroid dehydrogenase family. Mol Biol Evol 18 2154–2161 Occurrence Handle11719564
JR Brown CJ Douady MJ Italia WE Marshall MJ Stanhope (2001) ArticleTitleUniversal trees based on large combined protein sequence data sets. Nat Genet 28 281–285 Occurrence Handle10.1038/90129 Occurrence Handle1:CAS:528:DC%2BD3MXltFSmurw%3D Occurrence Handle11431701
K Bush GA Jacoby AA Medeiros (1995) ArticleTitleA functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39 1211–1233 Occurrence Handle1:CAS:528:DyaK2MXlvFOntLo%3D Occurrence Handle7574506
G Caetano-Anolles (2002) ArticleTitleTracing the evolution of RNA structure in ribosomes. Nucleic Acids Res 30 2575–2587 Occurrence Handle10.1093/nar/30.11.2575 Occurrence Handle12034847
A Carattoli (2001) ArticleTitleImportance of integrons in the diffusion of resistance. Vet Res 32 243–259 Occurrence Handle10.1051/vetres:2001122 Occurrence Handle11432416
D-F Feng G Cho RF Doolittle (1997) ArticleTitleDetermining the divergence times with a protein clock: Update and reevaluation. Proc Natl Acad Sci USA 94 13028–13033 Occurrence Handle10.1073/pnas.94.24.13028 Occurrence Handle9371794
M Galleni J Lamotte-Brasseur GM Rossolini J Spencer O Dideberg JM Frere (2001) ArticleTitleStandard numbering scheme for class B beta-lactamases. Antimicrob Agents Chemother 45 660–663 Occurrence Handle10.1128/AAC.45.3.660-663.2001 Occurrence Handle11181339
BG Hall (2001) Phylogenetic trees made easy: A how-to manual for molecular biologists. Sinauer Associates Sunderland, MA
BG Hall S Salipante M Barlow (2003) ArticleTitleThe metallo-β-lactamases fall into two distinct phylogenetic groups. J Mol Evol 57 249–254 Occurrence Handle1:STN:280:DC%2BD3MzjsF2qtA%3D%3D Occurrence Handle11403692
E Hannecart-Pokorni F Depuydt L de wit E van Bossuyt J Content R Vanhoof (1997) ArticleTitleCharacterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Im [corrected] associated with a sulI-type integron. Antimicrob Agents Chemother 41 314–318
JP Huelsenbeck F Ronquist (2001) ArticleTitleMrBayes: Bayesian inference of phylogeny. Bioinformatics 17 754–755 Occurrence Handle10.1093/bioinformatics/17.8.754 Occurrence Handle1:STN:280:DC%2BD3MvotV2isw%3D%3D Occurrence Handle11524383
B Jaurin T Grundstrom (1981) ArticleTitleAmpC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci USA 78 4897–4901 Occurrence Handle6795623
MS Johnson MJ Sutcliffe TL Blundell (1990) ArticleTitleMolecular anatomy: Phyletic relationships derived from three-dimensional structures of proteins. J Mol Evol 30 43–59 Occurrence Handle2107323
EV Koonin KS Makarova L Aravind (2001) ArticleTitleHorizontal gene transfer in prokaryotes: Quantification and classification. Annu Rev Microbiol 55 709–742 Occurrence Handle10.1146/annurev.micro.55.1.709 Occurrence Handle1:CAS:528:DC%2BD3MXnslKjtbw%3D Occurrence Handle11544372
B Mau M Newton (1997) ArticleTitlePhylogenetic inference for binary data on dendrograms using Markov chain Monte Carlo. J Comput Graph Stat 6 122–131
B Mau M Newton B Larget (1999) ArticleTitleBayesian phylogenetic inference via Markov chain Monte Carlo methods. Biometrics 55 1–12 Occurrence Handle1:STN:280:DC%2BD3M3ntV2qsw%3D%3D Occurrence Handle11318142
AA Medeiros (1997) ArticleTitleEvolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin Infect Dis 24 S19–S45 Occurrence Handle8994778
H Ochman AC Wilson (1987) ArticleTitleEvolution in bacteria: Evidence for a universal substitution rate in cellular genomes. J Mol Evol 26 74–86 Occurrence Handle3125340
H Ogawara (1993) ArticleTitlePhylogenetic tree and sequence similarity of beta-lactamases. Mol Phylogenet Evol 2 97–111 Occurrence Handle10.1006/mpev.1993.1010 Occurrence Handle8025724
M Ouellette L Bissonnette PH Roy (1987) ArticleTitlePrecise insertion of antibiotic resistance determinants into Tn21-like transposons: Nucleotide sequence of the OXA-1 beta-lactamase gene. Proc Natl Acad Sci USA 84 7378–7382 Occurrence Handle2823258
MA Ragan (2001) ArticleTitleDetection of lateral gene transfer among microbial genomes. Curr Opin Genet Dev 11 620–626 Occurrence Handle10.1016/S0959-437X(00)00244-6 Occurrence Handle1:CAS:528:DC%2BD3MXnslajt7k%3D Occurrence Handle11682304
B Rannala ZH Yang (1996) ArticleTitleProbability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J Mol Evol 43 304–311 Occurrence Handle1:CAS:528:DyaK28XlvVahur8%3D Occurrence Handle8703097
BA Rasmussen K Bush (1997) ArticleTitleCarbapenem-hydrolyzing beta-lactamases. Antimicrob Agents Chemother 41 223–232 Occurrence Handle9021171
GM Rossolini MA Condemi F Pantanella JD Docquier G Amicosante MC Thaller (2001) ArticleTitleMetallo-beta-lactamase producers in environmental microbiota: New molecular class B enzyme in Janthinobacterium lividum. Antimicrob Agents Chemother 45 837–844 Occurrence Handle10.1128/AAC.45.3.837-844.2001 Occurrence Handle11181369
N Saitou M Nei (1987) ArticleTitleThe neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 Occurrence Handle1:STN:280:BieC1cbgtVY%3D Occurrence Handle3447015
SJ Salipante BG Hall (2003) ArticleTitleDetermining the limits of the evolutionary potential of an antibiotic resistance gene. Mol Biol Evol 20 653–659 Occurrence Handle10.1093/molbev/msg074 Occurrence Handle12679553
KJ Shaw PN Rather FJ Sabatelli et al. (1992) ArticleTitleCharacterization of the chromosomal aac(6′)-Ic gene from Serratia marcescens. Antimicrob Agents Chemother 36 1447–1455 Occurrence Handle1354954
KJ Shaw PN Rather RS Hare GH Miller (1993) ArticleTitleMolecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57 138–163 Occurrence Handle1:CAS:528:DyaK3sXhvVKgtbk%3D Occurrence Handle8385262
DL Swofford (2000) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates Sunderland, MA
TA Tatusova TL Madden (1999) ArticleTitleBLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174 247–250 Occurrence Handle1:CAS:528:DyaK1MXjtlOlu74%3D Occurrence Handle10339815
HY Wu GH Miller MG Blanco RS Hare KJ Shaw (1997) ArticleTitleCloning and characterization of an aminoglycoside 6′-N-acetyltransferase gene from Citrobacter freundii which confers an altered resistance profile. Antimicrob Agents Chemother 41 2439–2447 Occurrence Handle9371347
Acknowledgements
This study was supported by Grant GM60761 from the National Institutes of Health. We are grateful to Reviewer 1 for insightful comments on an earlier version of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hall, B.G., Barlow, M. Structure-Based Phylogenies of the Serine β-Lactamases . J Mol Evol 57, 255–260 (2003). https://doi.org/10.1007/s00239-003-2473-y
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-003-2473-y