Abstract
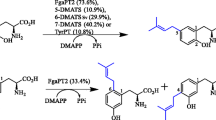
AuaA is a membrane-bound farnesyltransferase from the myxobacterium Stigmatella aurantiaca involved in the biosynthesis of aurachins. Like other known membrane-bound aromatic prenyltransferases, AuaA contains two conserved aspartate-rich motifs. Several amino acids in the first motif NXxxDxxxD were proposed to be responsible for prenyl diphosphate binding via metal ions like Mg2+. Site-directed mutagenesis experiments demonstrated in this study that asparagine, but not the arginine residue in NRxxDxxxD, is important for the enzyme activity of AuaA, differing from the importance of NQ or ND residues in the NQxxDxxxD or NDxxDxxxD motifs observed in some membrane-bound prenyltransferases. The second motif of known membrane-bound prenyltransferases was proposed to be involved in the binding of their aromatic substrates. KDIxDxEGD, also found in AuaA, had been previously speculated to be characteristic for binding of flavonoids or homogenisate. Site-directed mutagenesis experiments with AuaA showed that KDIxDxEGD was critical for the enzyme activity. However, this motif is very likely not specific for flavonoid or homogenisate prenyltransferases, because none of the tested flavonoids was accepted by AuaA or its mutant R53A in the presence of farnesyl, geranyl or dimethylallyl diphosphate.




Similar content being viewed by others
References
Akashi T, Sasaki K, Aoki T et al (2009) Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol 149:683–693
Ashby MN, Kutsunai SY, Ackerman S et al (1992) COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J Biol Chem 267:4128–4136
Brandt W, Brauer L, Gunnewich N et al (2009) Molecular and structural basis of metabolic diversity mediated by prenyldiphosphate converting enzymes. Phytochemistry 70:1758–1775
Bräuer L, Brandt W, Schulze D et al (2008) A structural model of the membrane-bound aromatic prenyltransferase UbiA from E. coli. Chembiochem 9:982–992
Haug-Schifferdecker E, Arican D, Brueckner R et al (2010) A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene dimethylallyltransferase reaction. J Biol Chem 285:16487–16494
Heide L (2009) Prenyl transfer to aromatic substrates: genetics and enzymology. Curr Opin Chem Biol 13:171–179
Jost M, Zocher G, Tarcz S et al (2010) Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity. J Am Chem Soc 132:17849–17858
Kaur J, Bachhawat AK (2009) A modified Western Blot protocol for enhanced sensitivity in the detectionof a membrane protein. Anal Biochem 384:348–349
Kuzuyama T, Noel JP, Richard SB (2005) Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435:983–987
Leviatan S, Sawada K, Moriyama Y et al (2010) Combinatorial method for overexpression of membrane proteins in Escherichia coli. J Biol Chem 285:23548–23556
Li S-M (2010) Prenylated indole derivatives from fungi: structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat Prod Rep 27:57–78
Melzer M, Heide L (1994) Characterization of polyprenyldiphosphate: 4-hydroxybenzoate polyprenyltransferase from Escherichia coli. Biochim Biophys Acta 1212:93–102
Metzger U, Schall C, Zocher G et al (2009) The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci USA 106:14309–14314
Metzger U, Keller S, Stevenson CE et al (2010) Structure and mechanism of the magnesium-independent aromatic prenyltransferase CloQ from the clorobiocin biosynthetic pathway. J Mol Biol 404:611–626
Ohara K, Muroya A, Fukushima N et al (2009) Functional characterization of LePGT1, a membrane-bound prenyltransferase involved in the geranylation of p-hydroxybenzoic acid. Biochem J 421:231–241
Pojer F, Wemakor E, Kammerer B et al (2003) CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci USA 100:2316–2321
Sasaki K, Mito K, Ohara K et al (2008) Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol 146:1075–1084
Sasaki K, Tsurumaru Y, Yamamoto H et al (2011) Molecular characterization of a membrane-bound prenyltransferase specific for isoflavone from Sophora flavescens. J Biol Chem 286:24125–24134
Stec E, Steffan N, Kremer A et al (2008) Two Lysine residues are responsible for the enzymatic activities of indole prenyltransferases from fungi. Chembiochem 9:2055–2058
Stec E, Pistorius D, Muller R et al (2011) AuaA, a membrane-bound farnesyltransferase from Stigmatella aurantiaca, catalyzes the prenylation of 2-methyl-4-hydroxyquinoline in the biosynthesis of aurachins. Chembiochem 12:1724–1730
Steffan N, Grundmann A, Yin W-B et al (2009) Indole prenyltransferases from fungi: a new enzyme group with high potential for the production of prenylated indole derivatives. Curr Med Chem 16:218–231
Yazaki K, Kunihisa M, Fujisaki T et al (2002) Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and characterization of a key enzyme in shikonin biosynthesis. J Biol Chem 277:6240–6246
Yazaki K, Sasaki K, Tsurumaru Y (2009) Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70:1739–1745
Yu X, Li S-M (2011) Prenylation of flavonoids by using a dimethylallyltryptophan synthase 7-DMATS from Aspergillus fumigatus. Chembiochem 12:2280–2283
Yu X, Xie X, Li S-M (2011) Substrate promiscuity of secondary metabolite enzymes: prenylation of hydroxynaphthalenes by fungal indole prenyltransferases. Appl Microbiol Biotechnol 92:737–748
Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to S.-M. Li).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Stec, E., Li, SM. Mutagenesis and biochemical studies on AuaA confirmed the importance of the two conserved aspartate-rich motifs and suggested difference in the amino acids for substrate binding in membrane-bound prenyltransferases. Arch Microbiol 194, 589–595 (2012). https://doi.org/10.1007/s00203-012-0795-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-012-0795-0