Abstract
Aims/hypothesis
Gestational diabetes mellitus (GDM) is associated with an increased risk of pre-eclampsia, macrosomia and the future development of type 2 diabetes mellitus in both mother and child. Although an early and accurate prediction of GDM is needed to allow intervention and improve perinatal outcome, no single protein biomarker has yet proven useful for this purpose. In the present study, we hypothesised that multimarker panels of serum proteins can improve first-trimester prediction of GDM among obese and non-obese women compared with single markers.
Methods
A nested case–control study was performed on first-trimester serum samples from 199 GDM cases and 208 controls, each divided into an obese group (BMI ≥27 kg/m2) and a non-obese group (BMI <27 kg/m2). Based on their biological relevance to GDM or type 2 diabetes mellitus or on their previously reported potential as biomarkers for these diseases, a number of proteins were selected for targeted nano-flow liquid chromatography (LC) MS analysis. This resulted in the development and validation of a 25-plex multiple reaction monitoring (MRM) MS assay.
Results
After false discovery rate correction, six proteins remained significantly different (p<0.05) between obese GDM patients (n=135) and BMI-matched controls (n=139). These included adiponectin, apolipoprotein M and apolipoprotein D. Multimarker models combining protein levels and clinical data were then constructed and evaluated by receiver operating characteristic (ROC) analysis. For the obese, non-obese and all GDM groups, these models achieved marginally higher AUCs compared with adiponectin alone.
Conclusions/interpretation
Multimarker models combining protein markers and clinical data have the potential to predict women at a high risk of developing GDM.
Similar content being viewed by others
Introduction
Gestational diabetes mellitus (GDM) is defined as glucose intolerance diagnosed during pregnancy; it is associated with an increased risk of obstetric and neonatal complications such as pre-eclampsia, caesarean section, macrosomia, shoulder dystocia, neonatal hypoglycaemia and the future development of type 2 diabetes mellitus in both mother and child [1–4]. GDM is a common complication of pregnancy, and its prevalence depends on the population and the diagnostic criteria applied. Obesity is a major risk factor for GDM, and obese pregnant women have an up to eight times higher risk of developing GDM compared with normal weight pregnant women [5].
In Denmark, pregnant women undergo a selective screening procedure for GDM based on risk factors; GDM diagnosis is based on a 75 g OGTT with a 2 h plasma glucose threshold of 9.0 mmol/l [6]. This threshold is in accordance with previous recommendations from the European Diabetic Pregnancy Study Group [7]. Recently, new diagnostic criteria for GDM were recommended by the International Association of Diabetes and Pregnancy Study Groups (IADPSG). These would lower the diagnostic 2 h threshold value of a 75 g OGTT to 8.5 mmol/l. Once implemented, this criterion is expected to greatly increase the prevalence of GDM not only in Denmark but also globally, thus challenging the health economy [8]. The IADPSG criteria are based on second trimester OGTT values and evidence of adverse perinatal outcomes. This recommendation suggests ruling out overt diabetes in early pregnancy by measuring fasting glucose (≥7.0 mmol/l), random plasma glucose (≥11.1 mmol/l on confirmation) or HbA1c (≥6.5% [≥48 mmol/mol]) levels. It was suggested that fasting glucose in the range of 5.1–6.9 mmol/l (which exceeds the IADPSG GDM criteria but does not meet the criteria for overt diabetes) should be considered as diagnostic of GDM. A drop in fasting glucose occurs in early pregnancy because of natural physiological changes [9]; hence, the use of this GDM diagnostic threshold may not identify the same individuals in early and later pregnancy. Furthermore, fasting glucose tests and/or OGTTs are time-consuming and elevated fasting glucose in early pregnancy is not highly predictive of later GDM development [10]. For these reasons, a biomarker assay that enables the early and accurate prediction of GDM is warranted. This could be an important tool in future screening strategies to ensure that women at the highest risk undergo a diagnostic OGTT at gestational week 24–28.
At present, no serum biomarker with clinical utility has been identified and the potential utility in GDM screening has been investigated for only a limited number of proteins. Adiponectin is by far the best-studied protein biomarker for GDM but lacks the sensitivity and specificity required for clinical use [11–16]. Other proteins such as sex hormone binding protein (SHBG), retinol-binding protein 4 (RBP-4), resistin and C-reactive protein (CRP) have also been investigated as potential biomarkers of GDM, but conclusions have been variable [14, 17–23].
The current study took a targeted proteomics candidate biomarker approach and used multiple reaction monitoring (MRM) MS to assess the potential of a number of both previously suggested and novel serum biomarkers for GDM in obese and non-obese pregnant women. Specifically, we hypothesised that multimarker panels of serum proteins could improve first-trimester prediction of GDM in obese and non-obese women. We developed a 25-plex MRM-MS assay for simultaneously measuring serum proteins using a minimal amount of sample material and with low analytical imprecision. Subsequent multivariate modelling of the data improved the AUC values obtained by receiver operating characteristic (ROC) analysis.
Methods
Samples and clinical data
All samples used in this nested case–control study were obtained from a biobank of ∼20,000 first-trimester non-fasting serum samples (stored at −80°C) collected from the routine screening for Down syndrome at Odense University Hospital [24]. All samples used in this study were taken between gestational week 8 + 1 and 13 + 6, as corrected by crown-rump length using astraia version 1.23.4.1 (astraia software, Munich, Germany). The Danish selective screening strategy for GDM includes a set of risk factors: BMI of ≥27 kg/m2, family history of diabetes and previous birth of a child with macrosomia. It is based on a large prospective study and has a sensitivity of 81% and a specificity of 65% [25]. Women with two or more of these risk factors or who have previously had GDM are offered an early diagnostic OGTT between gestational weeks 14 and 20 and again during week 27–30, whereas women with just one risk factor are only offered the late OGTT. In addition, glucosuria diagnosed at any gestational age (GA) also generates a referral for an OGTT. Altogether, 199 early pregnancy samples from women later diagnosed with GDM were identified in the biobank. Inclusion criteria were a singleton pregnancy and HbA1c values of <6.5% (48 mmol/mol) at the time of GDM diagnosis. In addition, 208 random samples from women matched by BMI and year of sampling served as controls. Samples were divided into two groups in accordance with current Danish GDM screening guidelines: BMI of ≥27 kg/m2 (obese group) and <27 kg/m2 (non-obese group). For statistical power calculations, see electronic supplementary material (ESM) Methods. Clinical data corresponding to all 407 samples were manually retrieved from patient medical records, entered into a database (SPSS version 21.0, IBM Danmark, Holte, Denmark) and analysed.
All aspects of the study were approved by the local ethics committee (S-20130092).
Sample preparation and MRM-MS analysis
Samples were prepared for MRM analysis essentially as described by Overgaard et al (for a detailed description, see ESM Methods) [26]. Briefly, serum samples were diluted 1:20 in 50 mmol/l ammonium bicarbonate and 15 μl samples were denatured, reduced, alkylated and trypsinised. Individually adjusted amounts of heavy isotope labelled standard peptides were added to each sample to achieve a ratio 1:1 with the endogenous light peptides; all peptides were then purified using Oasis HLB 10 mg cartridges (Waters Danmark, Hedehusene, Denmark). Samples were dried and reconstituted in 0.1% formic acid. For each sample, 1 μg was run on an Easy-nLC II nano liquid chromatography (LC) system using a C18 trapping column (length 2 cm; internal diameter 100 μm) for desalting and a C18 analytical column (length 10 cm, internal diameter 75 μm) for peptide separation (Thermo Scientific Danmark, Hvidovre, Denmark). Peptides were eluted with a three-step 60 min gradient of 0.1% formic acid in acetonitrile at a flow rate of 300 nl/min. Peptides were ionised using a Nanospray Flex ion source (Thermo Scientific) and analysed on a TSQ Vantage triple quadruple mass spectrometer (Thermo Scientific) in selected reaction monitoring mode.
For intra- and interassay CV calculations, pooled serum quality control samples were prepared in triplicate and analysed in duplicate on the LC tandem MS system. Standard curves (ESM Fig. 1) were made in triplicate for each peptide by adding different concentrations of heavy isotope labelled standard peptide to the same pooled serum. The spiked samples were finally processed according to the protocol for clinical samples. The lower limit of quantification (LLOQ) for each peptide was derived from ESM Fig. 1 (shown in ESM Table 1).
Immunological assays
Adiponectin and resistin levels were measured using the DRP300 and DRSN00 ELISA kits (R&D Systems, Abingdon, UK). Double testing for serum pregnancy-associated plasma protein A (PAPP-A) and free β chain human chorionic gonadotropin (β-hCG) was performed as part of the routine Down syndrome screening using an automated time-resolved fluoro-immunoassay (AutoDELFIA; PerkinElmer, Turku, Finland) on a 1235 AutoDELFIA analyser. Further details on immunological assays are provided in the ESM Methods.
Data analysis and statistics
SRM raw files were processed using Pinpoint 1.3 (Thermo Scientific). Data for each peptide represents the peak area ratio between the endogenous light peptide and the heavy isotope labelled spiked peptide. All data (clinical, MRM and ELISA) were transferred to an SPSS 21.0 (IBM) database and statistical analyses was done using SPSS 21.0 and Excel 2003 (Microsoft Danmark, Hellerup, Denmark).
Prior to the analysis, the dataset for each group (i.e. obese and non-obese) was randomised into a training set (comprising 75% of the data) to be used for biomarker verification and model development, and a validation set (comprising the remaining 25% of the data) to be used for model validation. Within each of the four data subsets, there was no significant difference in BMI between GDM patients and controls.
A significance level of p<0.05 was applied to all statistical tests used in this study.
Results
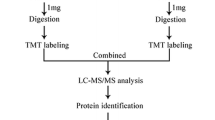
An overview of the sample collection, method development and data analysis procedures is provided in Fig. 1. Altogether, the levels of 29 proteins in 407 first-trimester serum samples were measured and analysed along with the relevant clinical data to investigate their potential as biomarkers (either individually or combined in multimarker models) for the early prediction of GDM.
Baseline clinical data
Clinical data for all patient groups are listed in Table 1. For both the obese and non-obese groups, women with GDM were significantly older than control participants. Women diagnosed with GDM also gave birth significantly earlier than those in the control group (p<0.001 for both groups). This difference is probably explained by the routine use of labour induction up to 2 weeks before term for women with GDM; in the obese group, this is reflected in a lower birthweight and a smaller birth length and abdominal circumference. After GA was taken into account, there was no significant difference in the number of large for GA (LGA; i.e. >90th percentile) children born to women with GDM and controls. In contrast, the frequency of pre-eclampsia and caesarean sections were significantly higher in the obese GDM group compared with obese controls (p = 0.032 and p = 0.039, respectively), whereas differences between the non-obese groups were not significant.
MRM assay development
We identified 54 candidate protein biomarkers from previous studies evaluating their potential as biomarkers for GDM and/or type 2 diabetes mellitus. Of these, 47 were selected for MRM-MS assay development because the characteristics of their trypsin-derived peptides (i.e. length, hydrophobicity, abundance and uniqueness) were theoretically compatible with the method. For a detailed description of the literature-based candidate search strategy and MRM assay peptide and transition selection, see ESM Fig. 2 and ESM Methods, respectively. The initial MRM-MS assay was tested on a pool of first-trimester serum and optimised using a spike of co-eluting heavy isotope labelled standard peptides to validate peptide identification and adjust for variations in sample preparation and LC-MS performance.
The final 25-plex MRM-MS assay measured the relative levels of 25 different proteins as represented by a total of 43 peptides (ESM Table 1). In this assay, 18 proteins were each represented by two peptides (for each protein, there was a high degree of correlation between peptides in the complete dataset; ESM Fig. 3) and seven proteins were each represented by a single peptide. Of the total of 25 proteins, 20 were represented by a minimum of five transitions. Standard curves verifying a linear correlation between amount of peptide and the intensity of the MS signal are shown in ESM Fig. 1. Peptide levels for all clinical samples were within the linear part of the standard curves and above the LLOQ shown in ESM Table 1. The interassay CVs for all peptides in the 25-plex MRM-MS assay were between 1.9% and 7.5% (ESM Table 1). Finally, the MRM assay was validated by a methods comparison analysis of three selected proteins for which either a clinical immunoassay or ELISA kit was available (ESM Fig. 4). The r values obtained for SHBG, apolipoprotein B (Apo B) and fibronectin were 0.97, 0.86 and 0.91, respectively.
Statistical analysis of the candidate biomarker panel
In addition to the 25 proteins measured by MRM-MS, levels of two adipokines (adiponectin and resistin) were measured by ELISA and serum concentrations for PAPP-A and β-hCG along with maternal age were retrieved from patient medical records (ESM Table 2). The potential of each protein to differentiate between GDM cases and controls was evaluated by applying the Mann–Whitney U test separately to the obese and non-obese training sets. For the obese group, nine variables were significantly different between GDM cases and controls and seven remained so after false discovery rate correction by the Benjamini–Hochberg procedure (Table 2). Of these, five were measured in the 25-plex MRM assay. For the non-obese group, three variables were significantly different between GDM cases and controls; however, none of these differences remained significant after correction. For all variables that initially showed significant differences between cases and controls, ROC curves were constructed and AUC values evaluated (Table 2 and Fig. 2a, b). For the obese group, adiponectin was the best independent predictor of GDM (AUC 0.712 [95% CI 0.642, 0.783]) followed by Apo M (AUC 0.633 [95% CI 0.557, 0.710]). For the non-obese group, adiponectin also showed the best predictive performance (AUC 0.671 [95% CI 0.563, 0.778]), followed by Apo D (AUC 0.659 [95% CI 0.551, 0.768]) and Apo L1 (AUC 0.634 [95% CI 0.524, 0.744]). These results indicate that although adiponectin is potentially a good universal predictor of GDM, apolipoproteins have distinct discriminative properties in subgroups of GDM patients.
ROC analysis of univariables and multivariate models. Univariate analysis of selected variables based on the training set of the (a) obese and (b) non-obese groups. Adiponectin (black line), Apo M (grey line in a) and Apo D (grey line in b). Multivariate analysis of (c) Model 2, obese group, (d) Model 7, non-obese group, (e) the core model obese group and the (f) core model non-obese group based on the training (black line) and validation (grey line) set. Red circles, the optimal cut-off points for high, medium and low specificity; dashed line; reference
Model development and validation
To further investigate the potential use of the biomarker panel evaluated in this study, multivariate models for predicting GDM were constructed using different combinations of variables (ESM Tables 3–6). All models were developed using binominal logistic regression analysis of a training set for each group (obese or non-obese) and then tested using a smaller validation set to rule out any overfitting of the data. For the same reason, no more than five variables were permitted for each model. Despite different modelling strategies, a number of variables recurred in several of the final models: namely maternal age adiponectin, Apo M and Apo L1 for the obese group; and Apo D and Apo L for the non-obese group. All models, including a 4-plex core panel common to both study groups, were evaluated by ROC analysis and corresponding AUC values (Table 3, Fig. 2c–f and ESM Table 3). Of the models listed in Table 3, Model 2 and Model 7 had the best overall predictive performance and showed the least difference in AUC values when tested in the validation set of each group (obese or non-obese). Moreover, both performed better than any of the single variables alone, including adiponectin (Table 2).
Based on the ROC analysis of Model 2 and Model 7, predictive values were calculated for high, medium and low specificity using the cohort prevalence of GDM in obese (2%) and non-obese (0.5%) pregnant women (Table 4). Similar calculations were also performed to account for an estimated future prevalence of 10% (expected after implementation of the new diagnostic IADPSG criteria for GDM). As shown in Table 4, the obese and non-obese models have a similar performance but different optimal cut-off points. As expected, positive predictive values (PPVs) increase with an increased prevalence of GDM. This is also evident for the estimated prevalence of 10%, assuming the models perform equally well for the new diagnostic threshold.
Discussion
In the present nested case–control study, we used MRM-MS and ELISA to identify and validate two 5-plex multimarker models that discriminate between: (1) GDM in obese pregnant women and BMI-matched obese control participants; and (2) GDM in non-obese pregnant women and BMI-matched non-obese control participants. Our data confirm that the adipocyte-derived hormone, adiponectin, is a specific independent predictor of GDM in both obese and non-obese women and directly compares its performance with those of a number of other previously suggested markers such as CRP, SHBG and the adipokines neutrophil gelatinase-associated lipocalin (NGAL), resistin and RBP-4 [14, 17–23, 27–32]. Of the novel candidate biomarkers included in our multiplex MRM-MS assay, Apo M, Apo D, Apo L1 and Apo AIV all appeared in the final multimarker models for predicting GDM.
In recent years, the MRM-MS method has gained immense popularity within the field of proteomics based on its application in biomarker development studies [33, 34]. Our study demonstrates how targeted MS applied to a classic nested case–control study, when adequately powered to detect subtle differences in serum protein levels, can support the development of multimarker models with better disease prediction. This approach also allowed us to directly compare a number of protein markers for which published results have been contradictory or inconclusive. A substantial advantage of MRM-MS assays is their compatibility with the often limited sample amount available in retrospective studies; our 25-plex MRM-MS assay uses less than 1 μl of crude serum. Although we used isotopically labelled peptide standards to correct for analytical and solid-phase extraction variances, the assay can potentially be affected by differences in trypsin digestion efficiencies. We chose a 21-h endpoint for the sample preparation protocol and validated the MRM assay by a methods comparison analysis of SHBG, Apo B and fibronectin; high r values were obtained for each (0.97, 0.86 and 0.91, respectively). As expected, the highest degree of correlation was obtained for the comparison between the MRM assay and an automated Siemens Immulite immunoassay. Apo B and fibronectin data were comparable between the MRM assay and manual ELISAs; for Apo B, the degree of correlation was similar to that previously obtained in a comparative study of LC-MS and nephelometry [35]. Taken together, we conclude that the MRM assay used in this study is fit for purpose but that further development of more focused marker panels with internal digestion controls would be valuable. A clear limitation of the method per se is its relatively low analytical sensitivity for serum or plasma (low μg/ml) when used without any prior reduction of sample complexity [26, 36]. Nevertheless, targeted MS offers an interesting analytical platform for future multimarker-based prenatal screening.
In this study, we divided the dataset into a training set for univariate and multivariate analyses and a separate validation set for testing the logistic prediction models and, thus, selection of those proteins with the best predictive performance. Of the 29 potential protein biomarkers, adiponectin is the best studied in relation to GDM [11–14, 16]. Here, adiponectin was the best performing independent predictor of GDM, regardless of BMI grouping. SHBG, RBP-4, CRP and resistin have also been described as potential biomarkers of GDM in numerous publications [14, 17–23, 28–31]. In this study, however, this potential was confirmed only for SHBG in obese women; in contrast, there was no significant difference in RBP-4, CRP and resistin levels between GDM patients and controls. This discrepancy may be attributable to sample collection at different GAs or statistical uncertainty due to a limited sample size.
The many apolipoproteins included in the 25-plex MRM-MS assay were primarily chosen based on their previously reported potential as biomarkers of GDM and/or type 2 diabetes mellitus (ESM Fig. 2) [37–39]. The best independent predictive apolipoproteins were Apo M, Apo D and Apo L1 (Tables 2 and 3); the performance of Apo M and Apo L1 depended on BMI grouping. After logistic regression modelling, Apo CIII emerged as a significant contributory variable in two obese models (Table 3). In contrast, Apo M and Apo L1 were included in both obese and non-obese models. This is therefore the first study to show that apolipoproteins have a predictive potential for GDM and a dependency on BMI.
Maternal age also proved to be significantly different between GDM cases and controls; thus, it was included in the final models (Table 3 and ESM Tables 3 and 4). No correlation was observed between maternal age and any of the protein markers included in the models. To evaluate the contribution of each marker to the AUC, we performed a ‘leave one out’ analysis (ESM Table 5). Apo M and adiponectin were the best single contributors in Model 2 (obese group), whereas Apo L1 made the greatest contribution to Model 7 (non-obese group). Based on these data, a ‘core’ panel for both groups was identified comprising adiponectin, Apo M, maternal age and Apo L1 (Table 3). The AUCs of the core panel were comparable with those of Model 2 and Model 7 when all data were analysed together regardless of BMI (all GDM group). In contrast, the core model performed marginally worse in the obese and non-obese groups. Taken together, the lack of enhanced discriminative power of each model in the obese and non-obese group compared with the all GDM group supports a universal rather than a BMI-stratified screening approach using different marker panels.
Although the predictive performance of both Model 2 (AUC 0.749) and Model 7 (AUC 0.739) was increased compared with the AUCs of single markers (Table 2), their performance in a low GDM prevalence population is only moderate. Moreover, although implementation of the IADPSG diagnostic criteria for GDM is expected to increase the prevalence to the vicinity of 10%, the associated test PPV of 19% for Model 2 at a medium cut-off point of 75% sensitivity and 64% specificity seems fair but not game changing. For a future screening regime, we envision the combined use of maternal history data, weight, age and biomarkers for risk calculation and, thus, stratification of the diagnostic OGTT. A similar multifactorial screening regime is widely used for the first-trimester detection of major aneuploidies including trisomy 21 [40], as well as for pre-eclampsia screening, which is currently being implemented at many prenatal screening clinics [41].
Clearly, the availability of complete maternal history data (i.e. previous GDM, previous birth of macrosomia child and diabetes in the family) for evaluating the selective screening regime both alone and in combination with serum markers would have strengthened this study. Furthermore, we used a nested case–control design, which can potentially lead to inflation of the AUC of the ROC compared with population-based studies. The study design has nevertheless been shown to be an efficient alternative to cross-sectional studies for the exploration of potential predictors [42]. We acknowledge that the ROC evaluation of the prediction models in the non-obese group was limited by sample size, although it still reached statistical significance. Clearly, GDM screening performance using the suggested models should be validated in prospective studies and should preferably also include maternal history data.
Another limitation of the current study was that only obese women in the control group underwent a diagnostic OGTT, whereas lean control women were not tested. Thus, we have no proof of normoglycaemia in pregnancy for the latter group. However, all women had urine glucose measured at every visit to the outpatient clinic throughout pregnancy, and any level of glucosuria would have resulted in a subsequent OGTT. We therefore consider GDM to be unlikely in the lean control women. The use of HbA1c measurement to rule out overt diabetes at the time of GDM diagnosis is another limitation to the study. HbA1c measurement forms part of our routine clinical analysis to identify women at risk of diabetic complications such as retinopathy and nephropathy. HbA1c levels decrease during pregnancy because of physiological changes [43]; thus, the cut-off level of 6.5% (48 mmol/mol) might be too high and so identify some women with more severe glucose intolerance.
In conclusion, this study has provided a comprehensive overview of the performance of protein biomarkers in early GDM prediction. Evidently, multivariate models using different combinations of predictors can improve the discriminative power over single markers.
Overall, the performance of the current models leaves room for additional metabolic and proteomic studies to identify novel early markers with even better discriminative power. Importantly, assays for novel candidate markers should detail molecular target specificity and be accessible to the research field. To this end, targeted MS offers an excellent platform for preclinical biomarker development to improve future prenatal care.
Abbreviations
- Apo:
-
Apolipoprotein
- CRP:
-
C-reactive protein
- GA:
-
Gestational age
- β-hCG:
-
Beta chain human chorionic gonadotropin
- GDM:
-
Gestational diabetes mellitus
- IADPSG:
-
International Association of Diabetes and Pregnancy Study Groups
- LC:
-
Liquid chromatography
- LGA:
-
Large for gestational age
- LLOQ:
-
Lower limit of quantification
- MS:
-
Mass spectrometry
- MRM:
-
Multiple reaction monitoring
- PAPP-A:
-
Pregnancy-associated plasma protein A
- PPV:
-
Positive predictive value
- RBP-4:
-
Retinol-binding protein 4
- ROC:
-
Receiver operating characteristic
- SHBG:
-
Sex hormone binding protein
References
Group HSCR, Metzger BE, Lowe LP et al (2008) Hyperglycemia and adverse pregnancy outcomes. N Engl J Med 358:1991–2002
Kim C, Newton KM, Knopp RH (2002) Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care 25:1862–1868
Ovesen PG, Jensen DM, Damm P, Rasmussen S, Kesmodel US (2015) Maternal and neonatal outcomes in pregnancies complicated by gestational diabetes. A nation-wide study. J Matern Fetal Neonatal Med: 1–5
Coustan DR (2013) Gestational diabetes mellitus. Clin Chem 59:1310–1321
Chu SY, Callaghan WM, Kim SY et al (2007) Maternal obesity and risk of gestational diabetes mellitus. Diabetes Care 30:2070–2076
Jensen DM, Damm P, Sorensen B et al (2003) Proposed diagnostic thresholds for gestational diabetes mellitus according to a 75-g oral glucose tolerance test. Maternal and perinatal outcomes in 3260 Danish women. Diabet Med 20:51–57
Lind T, Phillips PR (1991) Influence of pregnancy on the 75-g OGTT. A prospective multicenter study. The Diabetic Pregnancy Study Group of the European Association for the Study of Diabetes. Diabetes 40(Suppl 2):8–13
International Association of Diabetes and Pregnancy Study Groups Consensus Panel, Metzger BE, Gabbe SG et al (2010) International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 33:676–682
Mills JL, Jovanovic L, Knopp R et al (1998) Physiological reduction in fasting plasma glucose concentration in the first trimester of normal pregnancy: the diabetes in early pregnancy study. Metabolism 47:1140–1144
Corrado F, D’Anna R, Cannata ML, Interdonato ML, Pintaudi B, Di Benedetto A (2012) Correspondence between first-trimester fasting glycaemia, and oral glucose tolerance test in gestational diabetes diagnosis. Diabetes Metab 38:458–461
Ferreira AF, Rezende JC, Vaikousi E, Akolekar R, Nicolaides KH (2011) Maternal serum visfatin at 11–13 weeks of gestation in gestational diabetes mellitus. Clin Chem 57:609–613
Georgiou HM, Lappas M, Georgiou GM et al (2008) Screening for biomarkers predictive of gestational diabetes mellitus. Acta Diabetol 45:157–165
Lacroix M, Battista MC, Doyon M et al (2013) Lower adiponectin levels at first trimester of pregnancy are associated with increased insulin resistance and higher risk of developing gestational diabetes mellitus. Diabetes Care 36:1577–1583
Nanda S, Savvidou M, Syngelaki A, Akolekar R, Nicolaides KH (2011) Prediction of gestational diabetes mellitus by maternal factors and biomarkers at 11 to 13 weeks. Prenat Diagn 31:135–141
Ahnstrom J, Lindqvist PG, Walle U, Dahlback B (2010) Plasma levels of apolipoprotein M in normal and complicated pregnancy. Acta Obstet Gynecol Scand 89:1214–1217
Worda C, Leipold H, Gruber C, Kautzky-Willer A, Knofler M, Bancher-Todesca D (2004) Decreased plasma adiponectin concentrations in women with gestational diabetes mellitus. Am J Obstet Gynecol 191:2120–2124
Maged AM, Moety GA, Mostafa WA, Hamed DA (2014) Comparative study between different biomarkers for early prediction of gestational diabetes mellitus. J Matern Fetal Neonatal Med 27:1108–1112
Smirnakis KV, Plati A, Wolf M, Thadhani R, Ecker JL (2007) Predicting gestational diabetes: choosing the optimal early serum marker. Am J Obstet Gynecol 196:410, e1–6; discussion 410 e6–7
Caglar GS, Ozdemir ED, Cengiz SD, Demirtas S (2012) Sex-hormone-binding globulin early in pregnancy for the prediction of severe gestational diabetes mellitus and related complications. J Obstet Gynaecol Res 38:1286–1293
Su YX, Hong J, Yan Q et al (2010) Increased serum retinol-binding protein-4 levels in pregnant women with and without gestational diabetes mellitus. Diabetes Metab 36:470–475
Tepper BJ, Kim YK, Shete V, Shabrova E, Quadro L (2010) Serum retinol-binding protein 4 (RBP4) and retinol in a cohort of borderline obese women with and without gestational diabetes. Clin Biochem 43:320–323
Lewandowski KC, Stojanovic N, Bienkiewicz M et al (2008) Elevated concentrations of retinol-binding protein-4 (RBP-4) in gestational diabetes mellitus: negative correlation with soluble vascular cell adhesion molecule-1 (sVCAM-1). Gynecol Endocrinol 24:300–305
Qiu C, Sorensen TK, Luthy DA, Williams MA (2004) A prospective study of maternal serum C-reactive protein (CRP) concentrations and risk of gestational diabetes mellitus. Paediatr Perinat Epidemiol 18:377–384
Ekelund CK, Jorgensen FS, Petersen OB, Sundberg K, Tabor A, Danish Fetal Medicine Research Group (2008) Impact of a new national screening policy for Down’s syndrome in Denmark: population based cohort study. BMJ 337:a2547
Jensen DM, Molsted-Pedersen L, Beck-Nielsen H, Westergaard JG, Ovesen P, Damm P (2003) Screening for gestational diabetes mellitus by a model based on risk indicators: a prospective study. Am J Obstet Gynecol 189:1383–1388
Overgaard M, Cangemi C, Jensen ML, Argraves WS, Rasmussen LM (2015) Total and isoform-specific quantitative assessment of circulating fibulin-1 using selected reaction monitoring MS and time-resolved immunofluorometry. Proteomics Clin Appl 9:767–775
Lou Y, Wu C, Wu M, Xie C, Ren L (2014) The changes of neutrophil gelatinase-associated lipocalin in plasma and its expression in adipose tissue in pregnant women with gestational diabetes. Diabetes Res Clin Pract 104:136–142
Chen D, Fang Q, Chai Y, Wang H, Huang H, Dong M (2007) Serum resistin in gestational diabetes mellitus and early postpartum. Clin Endocrinol 67:208–211
Kuzmicki M, Telejko B, Szamatowicz J et al (2009) High resistin and interleukin-6 levels are associated with gestational diabetes mellitus. Gynecol Endocrinol 25:258–263
Noureldeen AF, Qusti SY, Al-Seeni MN, Bagais MH (2014) Maternal leptin, adiponectin, resistin, visfatin and tumor necrosis factor-alpha in normal and gestational diabetes. Indian J Clin Biochem 29:462–470
Vitoratos N, Deliveliotou A, Dimitrakaki A et al (2011) Maternal serum resistin concentrations in gestational diabetes mellitus and normal pregnancies. J Obstet Gynaecol Res 37:112–118
D’Anna R, Baviera G, Corrado F, Giordano D, Recupero S, Di Benedetto A (2009) First trimester serum neutrophil gelatinase-associated lipocalin in gestational diabetes. Diabet Med 26:1293–1295
Yassine H, Borges CR, Schaab MR et al (2013) Mass spectrometric immunoassay and MRM as targeted MS-based quantitative approaches in biomarker development: potential applications to cardiovascular disease and diabetes. Proteomics Clin Appl 7:528–540
Heegaard NH, Ostergaard O, Bahl JM et al (2015) Important options available–from start to finish–for translating proteomics results to clinical chemistry. Proteomics Clin Appl 9:235–252
Agger SA, Marney LC, Hoofnagle AN (2010) Simultaneous quantification of apolipoprotein A-I and apolipoprotein B by liquid-chromatography-multiple-reaction-monitoring mass spectrometry. Clin Chem 56:1804–1813
Kuzyk MA, Smith D, Yang J et al (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics 8:1860–1877
Kaur P, Rizk NM, Ibrahim S et al (2012) iTRAQ-based quantitative protein expression profiling and MRM verification of markers in type 2 diabetes. J Proteome Res 11:5527–5539
Li RX, Chen HB, Tu K et al (2008) Localized-statistical quantification of human serum proteome associated with type 2 diabetes. PLoS One 3, e3224
Kim SM, Park JS, Norwitz ER et al (2012) Identification of proteomic biomarkers in maternal plasma in the early second trimester that predict the subsequent development of gestational diabetes. Reprod Sci 19:202–209
Nicolaides KH (2011) Screening for fetal aneuploidies at 11 to 13 weeks. Prenat Diagn 31:7–15
Poon LC, Nicolaides KH (2014) Early prediction of preeclampsia. Obstet Gynecol Int 2014:297397
Biesheuvel CJ, Vergouwe Y, Oudega R, Hoes AW, Grobbee DE, Moons KG (2008) Advantages of the nested case–control design in diagnostic research. BMC Med Res Methodol 8:48
Nielsen LR, Ekbom P, Damm P et al (2004) HbA1c levels are significantly lower in early and late pregnancy. Diabetes Care 27:1200–1201
Acknowledgements
We thank V. Jørgensen, Odense University Hospital, Denmark, for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by a research grant from the Danish Diabetes Academy supported by the Novo Nordisk Foundation (to TR and DMJ) and by the Odense University Hospital Research Foundation (MO).
Duality of interest statement
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
All authors made substantive contributions to the study conception and design, data acquisition or data analysis and interpretation. All authors contributed to drafting the manuscript and approved the final version of the article. In addition, TR, DMJ, LMR and MO conceived the study; TR and MO developed the 25-plex MRM assay; TR performed the sample handling and ELISA analyses; TR and MO performed the statistical analyses; DMJ and LLTA identified patients and resolved clinical issues; and TR and NDT extracted data from the medical records. MO and DMJ are the guarantors of this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Methods
(PDF 161 kb)
ESM Fig. 1
(PDF 92 kb)
ESM Fig. 2
(PDF 121 kb)
ESM Fig. 3
(PDF 340 kb)
ESM Fig. 4
(PDF 94 kb)
ESM Table 1
(PDF 16 kb)
ESM Table 2
(PDF 19 kb)
ESM Table 3
(PDF 19 kb)
ESM Table 4
(PDF 21 kb)
ESM Table 5
(PDF 20 kb)
ESM Table 6
(PDF 28 kb)
Rights and permissions
About this article
Cite this article
Ravnsborg, T., Andersen, L.L.T., Trabjerg, N.D. et al. First-trimester multimarker prediction of gestational diabetes mellitus using targeted mass spectrometry. Diabetologia 59, 970–979 (2016). https://doi.org/10.1007/s00125-016-3869-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-016-3869-8