Abstract
Aims/hypothesis
Lipolytic breakdown of endogenous lipid pools in pancreatic beta cells contributes to glucose-stimulated insulin secretion (GSIS) and is thought to be mediated by acute activation of neutral lipases in the amplification pathway. Recently it has been shown in other cell types that endogenous lipid can be metabolised by autophagy, and this lipophagy is catalysed by lysosomal acid lipase (LAL). This study aimed to elucidate a role for LAL and lipophagy in pancreatic beta cells.
Methods
We employed pharmacological and/or genetic inhibition of autophagy and LAL in MIN6 cells and primary islets. Insulin secretion following inhibition was measured using RIA. Lipid accumulation was assessed by MS and confocal microscopy (to visualise lipid droplets) and autophagic flux was analysed by western blot.
Results
Insulin secretion was increased following chronic (≥8 h) inhibition of LAL. This was more pronounced with glucose than with non-nutrient stimuli and was accompanied by augmentation of neutral lipid species. Similarly, following inhibition of autophagy in MIN6 cells, the number of lipid droplets was increased and GSIS was potentiated. Inhibition of LAL or autophagy in primary islets also increased insulin secretion. This augmentation of GSIS following LAL or autophagy inhibition was dependent on the acute activation of neutral lipases.
Conclusions/interpretation
Our data suggest that lysosomal lipid degradation, using LAL and potentially lipophagy, contributes to neutral lipid turnover in beta cells. It also serves as a constitutive negative regulator of GSIS by depletion of substrate for the non-lysosomal neutral lipases that are activated acutely by glucose.
Similar content being viewed by others
Introduction
Glucose-stimulated insulin secretion (GSIS) comprises two separate but interconnected metabolic pathways in the beta cell: the initiation pathway, which is K+ ATP channel dependent and results in increased calcium influx [1, 2], and the amplification pathway, which potentiates insulin secretion independently of further changes in cytosolic calcium [3, 4]. The mechanisms underlying amplification are much less well understood than those of initiation [5–8] but the turnover of endogenous lipid stores, and consequent generation of a lipid signalling molecule, is one possibility [5, 6, 9, 10]. Evidence for this includes the inhibition of GSIS by deletion of neutral lipases, such as hormone-sensitive lipase (HSL) [11, 12] and adipose triglyceride lipase (ATGL) [13], or their pharmacological blockade using the pan lipase inhibitor orlistat [14]. In addition, at least one substrate of the neutral lipases, triacylglycerol (TG), is hydrolysed during GSIS, as witnessed by an acute glucose-stimulated, and orlistat-inhibited, release of glycerol from beta cells [9, 15, 16].
In contrast to the established roles of neutral lipases in insulin secretion, the potential involvement of lysosomal acid lipase (LAL) has largely been unaddressed. Historically, the chief function of this enzyme, as established in other cell types, was considered to be the breakdown of cholesteryl ester (CE) delivered to the cell via the LDL pathway [17, 18]. However, LAL has also been implicated in the turnover of endogenous lipid stores (lipid droplets) via the process of lipophagy, first established in hepatocytes in 2009 [19]. Lipophagy is a specific form of autophagy, the process by which macromolecules are surrounded by the formation of an autophagosome (APG) and then degraded by fusion of the APG with the lysosome. This process depends on a set of autophagy-related genes (ATGs), which are responsible for the production of the double-membrane APG structure [20, 21]. Of the associated proteins, only ATG8/microtubule-associated protein light chain 3 (LC3)-II, produced from lipidation of a precursor (LC3-I), is known to be associated with mature APGs and therefore serves as a marker for their formation [21]. Lipophagy is now recognised as an alternative method of lipid hydrolysis, whereby lipid droplets are surrounded by an APG and then delivered to the lysosome for degradation [19, 22]. It has been shown in macrophages that LAL is the enzyme responsible for the breakdown of TG and CE delivered to the lysosome via this route [23]. In liver, LAL/lipophagy makes a quantitatively important contribution to overall TG degradation, especially following high-fat feeding [24, 25].
This link between autophagy and lipid degradation led us to investigate whether a lysosomal pathway of lipid breakdown, in concert with neutral lipid hydrolysis, might contribute to the regulation of insulin secretion. In addressing these issues we now demonstrate for the first time that LAL and lipophagy are key negative regulators of a pool of neutral lipid in beta cells, which is mobilised as an amplification signal during GSIS.
Methods
Reagents
All tissue culture media, supplements and trypsin for MIN6 cells and isolated islets were from Gibco (Gaithersburg, MD, USA). Protease inhibitor tablets were from Roche Diagnostics (Penzburg, Germany). Insulin RIA kits were from Linco/Millipore (Billerica, NJ, USA). Phenylmethylsulfonyl fluoride (PMSF), sodium orthovanadate, DMSO, fatty acid free fraction V BSA and l-glucose, were from Sigma-Aldrich (St Louis, MO, USA). TaqMan reverse transcription kit and TaqMan PCR probes were from Applied Biosystems (Mulgrave, VIC, Australia). ATG7 (NM_028835) and LAL (NM_021460) ON-TARGETplus SMARTpool siRNA, control Non-Targeting siRNA and Dharmafect Transfection Reagent 3 were from Dharmacon (Pittsburgh, PA, USA). RNeasy kit was from Qiagen (Valencia, CA, USA). The BCA (bicinchoninic acid assay) protein assay kit was from Pierce (Rockford, IL, USA). The pre-cast NUPAGE gels, sample buffer, reducing agent, antioxidant and electrophoresis tank were from Invitrogen (Carlsbad, CA, USA). The transfer system for immunoblotting and the protein standard markers were from BioRad (Hercules, CA USA). The 96-well conical-bottom plates for islet insulin secretion assays were from Grenier Bio-one (Frickenhausen, Germany). Antibodies for immunoblotting/immunofluorescence were as follows: Anti-14-3-3 from Santa-Cruz (Santa Cruz, CA, USA), anti-ATG7 from Cell Signalling Technologies (Danvers, MA, USA), anti-LAL from Novus Biologicals (Littleton, CO, USA) and anti-LC3 from Sigma-Aldrich.
Cell culture and treatments
The pancreatic beta cell line MIN6 [26] was used at passages 26–35, as previously described [27]. Cells were grown at 37°C and 5% CO2 in DMEM (25 mmol/l glucose) supplemented with 10% FCS, 10 mmol/l HEPES, 50 U/ml of penicillin and 50 μg/ml streptomycin.
For Lalistat treatment, cells were seeded at 3 × 105 cells per well in 24-well plates or 4 × 105 cells per well in 12-well plates, and treated at varying doses and times as indicated, or with 1:2,000 DMSO as control.
For small-interfering RNA (siRNA) transfection MIN6 cells were seeded in 12-well plates, as above. LAL ON-TARGETplus SMARTpool siRNA, ATG7 ON-TARGETplus SMARTpool siRNA or control non-targeting siRNA were transfected using Dharmafect3 transfection reagent. siRNA constructs were present for 24 h and the media was then changed to complete DMEM (25 mmol/l glucose) and cells were incubated for a further 48 h. This achieved 70–80% transfection efficiency as per the manufacturer’s instructions.
Cell death was determined, as previously described [28], using a Cell Death Detection ELISA (Roche Diagnostics, NSW, Australia).
Islet isolation and insulin secretion assays
Islets were isolated from male C57Bl6 mice as previously described [16]. After pancreatic digestion, islets were purified and incubated overnight in RPMI 1640 medium (11 mmol/l glucose) supplemented with 10% FCS, 0.2 mmol/l glutamine, 10 mmol/l HEPES, 50 U/ml of penicillin and 50 μg/ml streptomycin. For inhibitor studies, islets were cultured for a further 48 h with 5 μmol/l Lalistat, 5 mmol/l 3-methyladenine (3MA) or vehicle control, and the media was changed every 24 h. Batches of five islets were picked, with at least six replicates per group, and insulin secretion assays were carried out as follows: islets or MIN6 cells were preincubated for 1 h in Krebs–Ringer HEPES buffer (KRHB) containing 0.1% (wt/vol.) BSA and 2 mmol/l glucose. They were then incubated for 1 h at 37°C with KRHB containing either 2 or 20 mmol/l glucose. An aliquot of the buffer was taken and insulin release was measured by RIA.
Confocal microscopy
MIN6 cells were seeded onto glass coverslips at 2 × 105 cells per well, and grown in DMEM (25 mmol/l glucose) for 24 h. For Nile Red staining, coverslips were washed three times with 1 ml PBS pre-warmed to 37°C. Cells were fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature. The PFA was removed, 100 ng/ml Nile Red in 37°C HBSS (Hanks’ buffered saline solution) was added and the cells were incubated in the dark for 15 min at room temperature. Staining solution was removed and the coverslips were washed three times with 1 ml PBS (37°C). Coverslips were mounted using Prolong Gold with DAPI (Molecular Probes, Carlsbad, CA, USA) and sealed with nail polish. Slides were left to dry in the dark for at least 3 h and then visualised using confocal microscopy. For LAL and BODIPY 493/503 (Molecular Probes) co-staining, following washing and fixation as above, coverslips were then blocked for 45 min with 3% (wt/vol.) BSA and incubated with anti-LAL antibody (1:1,000) overnight at 4°C. Cells were then washed three times with PBS and incubated with anti-rabbit Alexafluor 647 (Molecular Probes) and 1 μg/ml BODIPY 493/503 for 1 h at room temperature, protected from light. Coverslips were then mounted as described above.
Lysosomal function assays
MIN6 cells were treated for 24 h with Lalistat, bafilomycin A (BA) or chloroquine (CQ) and the following assays were performed.
Lysosomal pH was measured by staining with 1 μmol/l LysoSensor Yellow/Blue DND-160 (Invitrogen) for 5 min following treatments. The cells were then washed with warm medium and examined under a Leica DMI 6000 SP8 confocal microscope using 405 nm excitation wavelength with 450 or 530 nm emission filters. The 530:450 nm emission ratio was calculated using the software Photoshop CS4 (Adobe Systems Incorporated, San Jose, CA, USA).
Lysosomal proteolysis was measured using DQ-BSA (Molecular Probes), as per reference [29]. Briefly, cells were seeded onto coverslips, incubated overnight with DQ-BSA (10 μg/ml) and then treated with inhibitors (24 h). Cells were then washed in PBS, fixed with 2% PFA and mounted using Prolong Gold with DAPI (Molecular Probes). Fluorescent particles were visualised by confocal microscopy and quantified using Image J software (National Institute of Health, Bethesda, MD, USA).
RNA analysis
Total RNA was extracted from MIN6 cells using an RNeasy minikit and cDNA synthesis was performed on 1.5 μg of DNA, using the TaqMan reverse transcription kit. Real-time PCR was carried out on an HT-7900 PCR machine using TaqMan probes. The following probes were used: Lipe/Hsl (Mm00495359_m1), Pnpla2/Atgl (Mm00503040_m1), Pnpla3/Adpn (Mm00504420_m1), Lpl (Mm00434770_m1) and Lipa/Lal (Mm00498820). Analysis was carried out using the standard curve method and relative gene expression was normalised to Tbp (Mm00446971) as an endogenous control.
Western blotting
Protein extracts from MIN6 cells were resolved on a pre-cast 12% SDS-PAGE gel and transferred to polyvinylidene fluoride membrane. After blocking for 1 h in 5% skimmed milk, membranes were probed with antibodies overnight at 4°C (anti-ATG7, anti-LAL and anti-LC3) and normalised to a loading control (anti-pan 14-3-3). Band quantification was undertaken using ImageJ.
Lipidomic profiling using MS
Following 24 h control or Lalistat treatment MIN6 cells were pelleted in ice-cold PBS (two wells of a six-well plate per condition) and subjected to lipid MS as previously described [28, 30].
Statistical analysis
All data are expressed as mean ± SEM. All statistics were performed using GraphPad Prism5 software (GraphPad Software, La Jolla, CA, USA), and subjected to two-way, one-way or repeated measures ANOVA (with the Bonferroni post hoc test), paired Student’s (two-tailed) t test or unpaired Student’s (two-tailed) t test.
Results
LAL is expressed in pancreatic beta cells
Several lipases are expressed in beta cells [13, 16], although Hsl/Lipe and Atgl/Pnpla2 have generally been considered the most abundant. There is, however, residual lipase activity in mouse models of HSL and ATGL depletion, so additional enzymes may contribute to lipid hydrolysis in pancreatic beta cells [11, 13]. To investigate lipase expression in the mouse pancreatic beta cell line, MIN6, we carried out RT-PCR (Fig. 1a). Lal mRNA was present in MIN6 cells and LAL protein was also abundantly expressed in MIN6 cells, mouse islets and the human liver cell line HEPG2 (Fig. 1b). To determine whether this enzyme plays a functional role in beta cells we employed siRNA to knockdown LAL protein (Fig. 1c) by 28.8 ± 0.04% compared with control siRNA transfected cells (p < 0.05, one-sample t test). No increase in insulin secretion was observed when correcting for protein content (Fig. 1d), although we observed a significant decrease in total insulin content following Lal siRNA treatment (Fig. 1e). When this decrease was taken into consideration there was a significant increase in GSIS but not in basal insulin secretion (Fig. 1f).
Characterisation of LAL expression and function in pancreatic beta cells. (a) RT-PCR analysis of lipases in MIN6 cells. (b) Western blot analysis of MIN6 cells and mouse islets showing expression of LAL protein; HepG2 cells were included as a positive control for the antibody. (c) Representative western blot image for LAL protein and loading control, following transfection of a control pool of siRNA or Lal siRNA. (d) Insulin secretion from MIN6 cells, treated with control (white bars) or Lal (black bars) siRNA, after 2 or 20 mmol/l glucose stimulation (n = 4 independent experiments). (e) Insulin content of MIN6 cells following control (white bar) or Lal (black bar) siRNA treatment (n = 4 independent experiments; **p < 0.01 Student’s unpaired t test). (f) Insulin secretion from MIN6 cells following control (white bars) or Lal (black bars) siRNA treatment, corrected for total insulin content (n = 4 independent experiments; *p < 0.05 Student’s unpaired t test)
Inhibition of LAL increases insulin secretion
To characterise the role of LAL in beta cell function more comprehensively, and to avoid confounding effects of incomplete gene knockdown, we employed Lalistat, an inhibitor that is highly specific for LAL over neutral lipases [23, 31, 32]. Lalistat pretreatment for 24 h dose dependently increased insulin secretion from MIN6 cells (Fig. 2a) during a subsequent glucose challenge. However, Lalistat was without effect when only present for 2 h during the glucose stimulation (Fig. 2b). This contrasts with the function of neutral lipases, which regulate insulin secretion acutely [14]. From more extensive time course analyses we established that 5 μmol/l Lalistat was effective by 8 h, and that this effect was sustained during treatment for at least 48 h (Fig. 2c). There was a slight, but non-significant, increase in basal insulin release following Lalistat treatment (Fig. 2a, c). Insulin content was unaffected at low Lalistat doses of 1 and 5 μmol/l although there was a non-significant increase observed at 10 μmol/l (Fig. 2d). Therefore the 5 μmol/l concentration was used thereafter to avoid content issues.
Inhibition of LAL using Lalistat increases insulin secretion. (a) MIN6 cells were treated for 24 h with 1, 5 or 10 μmol/l Lalistat. Insulin secretion over 1 h in response to 2 mmol/l (white bars) or 20 mmol/l (black bars) glucose is shown (n = 3 experiments; *p < 0.05 one-way ANOVA with Bonferroni post hoc test). (b) Insulin secretion from MIN6 cells treated for 2 h with 5 μmol/l Lalistat and 2 mmol/l glucose (white bars) or 20 mmol/l glucose (black bars) (n = 3 experiments). (c) Time course of Lalistat treatment; insulin secretion in response to 2 mmol/l glucose (white bars) and 20 mmol/l glucose (black bars) is shown (n = 4 experiments; *p < 0.05, **p < 0.01, Student’s paired t test). (d) Insulin content of MIN6 cells following various Lalistat doses (n = 3 experiments). (e) Insulin secretion in response to glucose, KCl, KCl + DZX and KCl + FSK after 24 h 5 μmol/l Lalistat (grey bars) or control treatment (white bars) (n = 4 experiments; *p < 0.05, **p < 0.0,1 repeated measures ANOVA with Bonferroni post hoc test). DZX, diazoxide; FSK, forskolin
To investigate the amplification pathway [3, 4], MIN6 cells were stimulated in the combined presence of diazoxide (to circumvent metabolic closure of K+ ATP channels) and KCl (to provide an independent depolarisation stimulus). Under these conditions, raising glucose from 2 to 20 mmol/l doubled insulin secretion, indicative of the amplification pathway (Fig. 2e). Lalistat pretreatment slightly augmented the response to KCl plus diazoxide at low glucose concentration but this was significantly further increased at high glucose concentration. Thus chronic LAL inhibition augments the amplification phase of GSIS. We next employed forskolin, which can also potentiate Ca2+-dependent secretion. Following Lalistat pretreatment, however, the increment in secretion, comparing KCl plus forskolin with KCl alone, was not statistically significant and less marked than the corresponding effect of 20 mmol/l glucose (Fig. 2e). This is consistent with the mechanism of action of forskolin (raising cAMP), which is not thought to contribute to the amplification pathway employed by glucose [3, 4]. Collectively these results suggest that LAL inhibition has a greater impact on the secretory response to glucose than non-nutrient secretagogues, and acts in the amplification pathway.
Lalistat does not adversely affect general lysosomal function in MIN6 cells
To check whether chronic LAL inhibition affects general lysosomal function, we employed BA and CQ, which disrupt the lysosomal pH gradient. Using LysoSensor pH probes, BA reduced the yellow/blue ratio, corresponding to the expected decrease in lysosomal acidity, but this was not observed after Lalistat treatment (Fig. 3a). Interestingly, Lalistat caused a significant increase in the total fluorescence emission following LysoSensor administration, which was not observed with BA treatment (Fig. 3b), indicative of an increase in total lysosome number. To assess whether Lalistat altered lysosomal protein degradation, cells were incubated with a fluorescently labelled BSA molecule (DQ-BSA), which when cleaved by proteolytic enzymes resident in the lysosome generates individual fluorescent particles that can be quantified to assess proteolysis. Lalistat treatment did not inhibit proteolytic activity but rather appeared to slightly increase the number of particles (Fig. 3c). This increase is most likely due to the increased lysosomal number (seen in Fig. 3b). Together, these results suggest that Lalistat does not cause general lysosomal disruption but rather specifically impacts on the lipid degradative capacity of the lysosome, and seemingly increases lysosomal number.
Lalistat treatment does not adversely affect lysosomal function. MIN6 cells were treated for 24 h with or without 5 μmol/l Lalistat, or overnight with BA (5 nmol/l) or CQ (5 μmol/l). (a) Lysosomal acidity was measured using LysoSensor (n = 3 independent experiments, visualising 26 fields of view; **p < 0.01, one-way ANOVA, Bonferroni post hoc test). AU, arbitrary units. (b) Total blue and yellow fluorescence from the LysoSensor assay was analysed (n = 3 independent experiments, visualising 26 fields of view per condition; ***p < 0.001 one-way ANOVA, Bonferroni post hoc test). (c) General proteosomal degradation was measured using fluorescently labelled DQ-BSA (n = 3 independent experiments, visualising 93–152 cells per condition: *p < 0.05 one-way ANOVA, Bonferroni post hoc test). (d) Insulin secretion results following BA (0.5 or 5 nmol/l) and CQ (5 or 50 μmol/l) treatment with 2 mmol/l (white bars) or 20 mmol/l (black bars) glucose stimulation (n = 3 experiments). (e) Insulin content of BA- or CQ-treated MIN6 cells (n = 3 experiments): *p < 0.05, Student’s unpaired t test. (f) Apoptosis following Lalistat, CQ or BA treatment. Results are expressed as fold change vs no treatment, corrected for DNA content (n = 3 experiments; *p < 0.05 one-sample t test)
We also measured insulin secretion from MIN6 cells in response to these general lysosomal inhibitors. Treatment with CQ completely abolished GSIS from MIN6 cells (Fig. 3d), but increasing doses of BA were without effect. There was also decreased insulin content following high-dose BA or CQ treatment (Fig. 3e) in contrast to Lalistat treatment, which had no effect or increased effect at high doses (Fig. 2d). Additionally, chronic Lalistat treatment had no effect on apoptosis in MIN6 cells, as measured by DNA fragmentation ELISA (Fig. 3f), whereas CQ treatment significantly increased apoptosis compared with no-treatment controls. Taken together, these data indicate that a general perturbation of lysosomal function is neither necessary nor sufficient for augmenting GSIS, suggesting that the effect of Lalistat is mediated more specifically by inhibition of LAL.
Lalistat increases the neutral lipid pool of MIN6 cells
We next determined the effect of LAL inhibition on lipid pools within MIN6 cells. Lipidomic analysis was carried out on cells treated with Lalistat for 24 h. Inhibiting LAL activity had no effect on total ceramide, monohexosyl ceramide or sphingomyelin levels (Fig. 4a) or on major phospholipids (Fig. 4b). Total levels of the neutral lipids, TG, diacylglyerol and CE were, however, all significantly increased following pretreatment (Fig. 4c).
Inhibition of LAL causes changes to the lipid profile of MIN6 cells. MIN6 cells were treated for 24 h with 5 μmol/l Lalistat (black bars) or control DMSO (white bars). The lipids were extracted and then subjected to MS, as described in the Methods. Results are expressed as mmol lipid per mg protein (n = 4 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, Student’s unpaired t test). (a) Total sphingolipids: amount of total sphingomyelin (SM), ceramide (Cer) and monohexosyl ceramide (MHC). (b) Total phospholipids: amount of total phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI) and phosphatidylserine (PS). (c) Total neutral lipids: amount of total diacylglycerol (DG), TG, CE and non-esterified cholesterol (COH)
Autophagy inhibition leads to increased insulin secretion
Because in other cell types, LAL has been implicated in the process of lipophagy [23], we next assessed whether inhibiting autophagy might recapitulate the augmentation of insulin secretion seen with Lalistat. We therefore knocked down ATG7 protein, which is an important initiator of APG formation, in MIN6 cells (Fig. 5a). This was accompanied by reduced accumulation of the downstream effector protein LC3-II (Fig. 5b), which reflects decreased APG formation compared with control siRNA-treated cells. Under these treatment conditions, inhibition of autophagy significantly increased GSIS from MIN6 cells (Fig. 5c), similar to chronic exposure to Lalistat. Knockdown of ATG7 caused a significant increase in total insulin content in MIN6 cells (Fig. 5d), which was not necessarily unexpected when inhibiting protein turnover by decreasing autophagy. However, when insulin secretion was corrected for total insulin content there was still a significant increase in GSIS from Atg7 siRNA-treated cells compared with control siRNA-treated cells (Fig. 5e).
Atg7 siRNA decreases autophagy and increases insulin secretion from MIN6 cells. MIN6 cells were transfected with control siRNA or Atg7 siRNA and left to culture for a total of 72 h. (a) Representative western blot image showing ATG7 and LC3 protein levels following control siRNA or Atg7 siRNA treatment. (b) Quantification of western blots for ATG7 and LC3-II protein for control siRNA (white bars) or Atg7 siRNA (black bars) treated MIN6 cells (n = 3 or 4 experiments; *p < 0.05, one-sample t test). (c) Insulin secretion following control siRNA (white bars) or Atg7 siRNA (black bars) treatment with 2 or 20 mmol/l glucose (n = 5 independent experiments; *p < 0.05, Student’s paired t test). (d) Insulin content after control (white bar) or Atg7 siRNA (black bar) treatment (n = 5 independent experiments; *p < 0.05, Student’s paired t test). (e) Insulin secretion following control (white bars) or Atg7 siRNA (black bars) treatment, corrected for insulin content (n = 4 independent experiments; *p < 0.05, Student’s paired t test)
We next investigated any potential feedback of LAL inhibition on autophagy. Under steady-state conditions, Lalistat treatment had no effect on total ATG7, LAL or LC3-II : LC-I ratio (Fig. 6a, b). Autophagic flux was also evaluated using MIN6 cells pre-exposed to Lalistat for 24 h and then acutely treated with CQ to inhibit downstream degradation of LC3-II (Fig. 6c, d). Under control conditions CQ significantly increased the LC3-II to LC3-I ratio, as expected, but this was attenuated by Lalistat. These results suggest some crosstalk between LAL inhibition and autophagy, albeit not complete.
There is crosstalk between LAL inhibition and autophagy. MIN6 cells were treated for 24 h with 5 μmol/l Lalistat or control and subjected to western blot analysis. (a) Representative western blot for autophagy markers (ATG7 and LC3-II) and LAL following Lalistat treatment. (b) Quantification of western blot results following control (white bars) or Lalistat (grey bars) treatment, for LC3-II:LC3-I ratio, ATG7 and LAL (n = 3 independent experiments). (c) Representative western blot for LC3 following 24 h Lalistat treatment and with or without acute (2 h) CQ addition. (d) Quantification of western blot results showing LC3-II : LCI ratio following 24 h Lalistat treatment with (black bars) or without (white bars) acute (2 h) CQ addition (n = 4 independent experiments; *p < 0.05, **p < 0.01 Student’s paired t test)
Inhibition of LAL and autophagy promotes lipid-droplet accumulation
Our lipidomic results (Fig. 4c) demonstrated a selective increase in neutral lipids following Lalistat treatment, and these lipids would be expected to be stored in pancreatic beta cells as lipid droplets [33–35]. Thus untreated MIN6 cells contained relatively few lipid droplets, but these were increased following 24 h Lalistat treatment (Fig. 7a–c). Moreover, inhibition of autophagy by knocking down ATG7 increased neutral lipid storage (Fig. 7d–f). Compared with control siRNA-treated cells, Atg7 siRNA increased lipid droplets approximately twofold (Fig. 7f). This indicates that inhibiting lysosomal lipid degradation, by blocking either LAL or autophagy, increases neutral lipid storage.
Lalistat and Atg7 siRNA increase lipid droplets. MIN6 cells were stained with Nile Red following either control (a), 24 h Lalistat (b), 72 h control siRNA (d) or 72 h Atg7 siRNA (e) to visualise lipid droplets (red staining); nuclei were stained with DAPI (blue). (c, f) Quantification of lipid droplets after control or Lalistat (c) (n = 3 independent experiments, visualising 73–99 cells per condition) or following siRNA treatment (f) (n = 4 independent experiments, visualising 58–66 cells per condition; *p < 0.05, Student’s unpaired t test). (g, h) MIN6 cells were also stained with BODIPY 493/503 to visualise neutral lipid (green) and LAL to visualise lysosomes (red) to determine co-localisation between neutral lipid storage and lysosomes in untreated (g) and Lalistat treated (h) cells. Scale bar, 10 μm
We also investigated the site of this lipid accumulation by comparing staining of LAL (as a marker of lysosomes) and BODIPY 493/503 (to stain neutral lipid). Under control conditions the few observed lipid bodies (green staining, Fig. 7g), did not localise with lysosomes (in red). Upon Lalistat treatment, the number of lipid bodies increased, but they remained outside of the lysosome (Fig. 7h), consistent with cytosolic lipid droplets.
Inhibition of LAL and autophagy in primary islets also increases insulin secretion
We next sought to confirm the roles of LAL and autophagy in regulating GSIS in primary beta cells. Pretreatment of isolated mouse islets with Lalistat for 48 h also caused a significant increase in insulin secretion (Fig. 8a), with no effect on total insulin content (Fig. 8b). Moreover, this increased GSIS due to chronic exposure to Lalistat was completely abolished when orlistat (a pan lipase inhibitor) was present acutely during the glucose stimulation. We next demonstrated that inhibiting autophagy in isolated mouse islets, using the chemical inhibitor 3MA, also increased insulin secretion (Fig. 8c), despite reducing insulin content (Fig. 8d). Interestingly, the increment in GSIS due to 3MA was also partially inhibited by orlistat (Fig. 8c). These results together suggest that inhibiting LAL or autophagy augments insulin secretion via a similar mechanism, most likely by augmenting a neutral lipid pool, which is acutely mobilised during GSIS via an orlistat-sensitive lipase.
Lalistat and inhibition of autophagy increase insulin secretion in primary mouse islets. (a) Mouse islets were isolated and cultured for 48 h in RPMI or RPMI + Lalistat. Insulin secretion was measured over 1 h in the presence of either 2 mmol/l (white bars) or 20 mmol/l (black bars) glucose, or 20 mmol/l glucose + 0.2 mmol/l orlistat (grey bars) (n = 9–12; *p < 0.05 Student’s paired t test). (b) Insulin content of islets following treatment for 48 h with Lalistat. (n = 9–12). (c) Mouse islets were isolated and cultured in the presence or absence of 5 mmol/l 3MA for 24 h. Insulin secretion was measured over 1 h in the presence of either 2 mmol/l (white bars) or 20 mmol/l (black bars) glucose, or 20 mmol/l glucose + 0.2 mmol/l orlistat (grey bars) (n = 6; *p < 0.05, ****p < 0.0001, one-way ANOVA, Bonferroni’s post hoc test). (d) Insulin content of islets treated with 3MA (n = 6); *p < 0.05, Student’s unpaired t test)
Discussion
Lipophagy, a specialised form of autophagy in which lipid droplets are degraded via lysosomal fusion, has been recently described in several tissues [19, 23, 36]. It appears to function as a means of mobilising lipid stores in times of nutrient demand [37], or as a means of helping to regulate cellular cholesterol homeostasis [23]. LAL has recently been implicated as the important metabolising enzyme in this process [23, 38]. We now provide the first evidence of a functional role for lipophagy/LAL in beta cells, by employing pharmacological and/or genetic inhibition of autophagy and LAL, in clonal MIN6 beta cells and isolated islets, and monitoring effects on insulin secretion, neutral lipid accumulation and autophagic markers. The generally excellent concordance between all these approaches suggests that lipophagy serves as a constitutive negative regulator of GSIS. It will be important in future studies to address how this role intersects with the better-known function of LAL in hydrolysing lipid from LDL [17, 18].
Although the contribution of the amplification pathway to GSIS is now well established, the underlying mechanisms remain obscure. One possibility focuses on the turnover of intracellular lipid stores [10, 39]. Evidence for this comes from studies in which lipase activity is inhibited either pharmacologically [14] or in HSL [11, 40] and ATGL [13] knockout mice, which show aberrant insulin secretion. Thus, while there is a key role for neutral lipases in GSIS, our study provides the first evidence that lysosomal lipid metabolism contributes as well, but in a very different manner. First, and most notably, inhibition of LAL potentiates GSIS rather than inhibiting it. Second, whereas neutral lipase inhibitors such as orlistat block secretion acutely, Lalistat required >8 h to exert its effects. Thus LAL does not appear to generate lipid signalling molecules itself, unlike neutral lipases. Third, the increment in GSIS due to chronic exposure to Lalistat was itself blocked by acute treatment with orlistat, suggesting that LAL acts upstream of the neutral lipases. The simplest explanation for these collective findings is that inhibition of LAL leads to slow accumulation of a neutral lipid signalling pool, which is subsequently mobilised by orlistat-sensitive lipases in response to glucose stimulation.
We also provide some evidence of potential crosstalk between LAL inhibition and autophagy. Chronic Lalistat treatment inhibited autophagic flux measured in the presence of CQ (Fig. 6d). This could help explain how inhibition of a lysosomal enzyme, LAL, might lead to a build up of neutral lipid stores, which we observed in non-lysosomal sites (presumably cytosol) where they can be accessed by neutral lipases during GSIS. However, there are other possible explanations, with very recent studies pointing to a complex inter-regulation of more traditional lipid metabolism with LAL/lipophagy mediated by transcription factors, such as TFEB/HLH-30 and MXL-3 [41, 42]. The exact mechanism for interaction between LAL and neutral lipid droplets in beta cells will require extensive further studies to resolve.
Although we provide evidence supporting a pivotal role for lipid metabolism and signalling in regulating GSIS, the exact identity of the signalling metabolite(s) responsible for this remains obscure. Candidates include long-chain fatty acyl CoAs, perhaps via protein acylation or modulation of ion channels [43], and diacylglycerol, albeit most probably acting via mechanisms independent of protein kinase C [44, 45]. These metabolites can be derived from breakdown of TG. However, our results suggest that turnover of CE should also be investigated in future studies, since prior augmentation of this pool correlated with increased insulin secretion (Fig. 4c). Moreover, the fact that both inhibition of LAL and autophagy have very similar effects in terms of potentiating GSIS and accumulation of lipid droplets points to a major role for LAL in mediating lipophagy. This is strongly underscored by the observation that potentiation of GSIS due to prior disruption of either LAL (by Lalistat) or autophagy (by 3MA) was sensitive to inhibition by orlistat, suggestive of a common upstream mechanism. However, this sensitivity was only partial in the case of 3MA, pointing either to LAL-independent effects of autophagy that contribute to GSIS, or LAL working through other mechanisms in addition to lipophagy. The latter would be consistent with the role of LAL in mobilising lipids bound to lipoprotein [17, 18].
The beneficial effects of inhibiting autophagy observed in our study (enhanced GSIS) contrasts with some recent studies utilising beta cell-specific ATG7 knockout mice, in which loss of autophagy impairs glucose tolerance [46, 47]. It must be stressed that our system employs a relatively mild and transient inhibition of autophagy, using siRNA, in contrast to the life-long and complete deletion of Atg7, which is characterised by reduced beta cell mass [46, 47]. Therefore, long-term inhibition of autophagy is detrimental to general beta cell function due to changes in protein, lipid and organelle degradation. In our study, we highlight the importance of specifically targeting one arm of these autophagic processes—lipid degradation. Also, there is some prior support for autophagy as a (short-term) negative regulator of beta cell responses, in that activation of autophagy by rapamycin resulted in decreased insulin secretion [48].
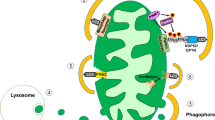
In conclusion, this study provides the first evidence that lipophagy/lysosomal lipid degradation contributes to neutral lipid metabolism in beta cells, as it does in other cell types (Fig. 9a). More importantly, our results suggest that lipophagy and LAL (lysosomal lipid degradation) serve as negative regulators of insulin secretion, and under normal conditions limit the size of the lipid signalling pool. Although it remains to be determined whether dysregulation of these processes contributes to beta cell failure in any way, their inhibition might be a beneficial means for pharmacologically augmenting insulin secretion (Fig. 9b).
Proposed model for lipophagy in beta cells. (a) Lipophagy influences insulin secretion by altering lipid metabolism in beta cells, for requirements during cellular processes. (b) Inhibition of autophagy or LAL causes a decreased turnover of neutral lipid pools, which are then acted upon acutely by neutral lipases (such as HSL or ATGL) upon glucose stimulation, leading to enhanced insulin release
Abbreviations
- APG:
-
Autophagosome
- ATG:
-
Autophagy-related genes
- ATGL:
-
Adipose triglyceride lipase
- BA:
-
Bafilomycin A
- BCA:
-
Bicinchoninic acid
- CE:
-
Cholesteryl ester
- CQ:
-
Chloroquine
- GSIS:
-
Glucose-stimulated insulin secretion
- HBSS:
-
Hanks’ buffered saline solution
- HSL:
-
Hormone-sensitive lipase
- KRHB:
-
Krebs–Ringer HEPES buffer
- LAL:
-
Lysosomal acid lipase
- LC3:
-
Microtubule-associated protein light chain 3
- 3MA:
-
3-Methyladenine
- PFA:
-
Paraformaldehyde
- siRNA:
-
Small-interfering RNA
- TG:
-
Triacylglycerol
References
Prentki M, Tornheim K, Corkey BE (1997) Signal transduction mechanisms in nutrient-induced insulin secretion. Diabetologia 40(suppl 2):S32–S41
Ashcroft F, Rorsman P (1990) ATP-sensitive K+ channels: a link between β-cell metabolism and insulin secretion. Biochem Soc Trans 18:109–111
Henquin JC (2000) Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes 49:1751–1760
Henquin JC, Dufrane D, Nenquin M (2006) Nutrient control of insulin secretion in isolated normal human islets. Diabetes 55:3470–3477
Prentki M, Vischer S, Glennon MC, Regazzi R, Deeney JT, Corkey BE (1992) Malonyl-CoA and long chain acyl-CoA esters as metabolic coupling factors in nutrient-induced insulin secretion. J Biol Chem 267:5802–5810
Nolan CJ, Prentki M (2008) The islet beta-cell: fuel responsive and vulnerable. Trends in Endocrinol Metab 19:285–291
MacDonald MJ, Fahien LA, Brown LJ, Hasan NM, Buss JD, Kendrick MA (2005) Perspective: emerging evidence for signaling roles of mitochondrial anaplerotic products in insulin secretion. Am J Physiol Endocrinol Metab 288:E1–E15
Jensen MV, Joseph JW, Ronnebaum SM, Burgess SC, Sherry AD, Newgard CB (2008) Metabolic cycling in control of glucose-stimulated insulin secretion. Am J Physiol Endocrinol Metab 295:E1287–E1297
Prentki M, Madiraju SRM (2012) Glycerolipid/free fatty acid cycle and islet β-cell function in health, obesity and diabetes. Mol Cell Endocrinol 353:88–100
Fex M, Mulder H (2008) Lipases in the pancreatic beta-cell: implications for insulin secretion. Biochem Soc Trans 36:885–890
Fex M, Haemmerle G, Wierup N et al (2009) A beta cell-specific knockout of hormone-sensitive lipase in mice results in hyperglycaemia and disruption of exocytosis. Diabetologia 52:271–280
Rl R, Masiello P, Wang SP, Li H, Mitchell GA, Prentki M (2001) A role for hormone-sensitive lipase in glucose-stimulated insulin secretion. Diabetes 50:1970–1975
Peyot M-L, Guay C, Latour MG et al (2009) Adipose triglyceride lipase is implicated in fuel- and non-fuel-stimulated insulin secretion. J Biol Chem 284:16848–16859
Mulder H, Yang S, Winzell MS, Holm C, Ahren B (2004) Inhibition of lipase activity and lipolysis in rat islets reduces insulin secretion. Diabetes 53:122–128
Cantley J, Biden TJ (2010) Targeting triglyceride/fatty acid cycling in beta-cells as a therapy for augmenting glucose-stimulated insulin secretion. Islets 2:127–129
Cantley J, Burchfield JG, Pearson GL, Schmitz-Peiffer C, Leitges M, Biden TJ (2009) Deletion of PKCε selectively enhances the amplifying pathways of glucose-stimulated insulin secretion via increased lipolysis in mouse beta-cells. Diabetes 58:1826–1834
Jelinek D, Patrick SM, Kitt KN, Chan T, Francis GA, Garver WS (2009) Physiological and coordinate downregulation of the NPC1 and NPC2 genes are associated with the sequestration of LDL-derived cholesterol within endocytic compartments. J Cell Biochem 108:1102–1116
Miller WL, Bose HS (2011) Early steps in steroidogenesis: intracellular cholesterol trafficking. J Lipid Res 52:2111–2135
Singh R, Kaushik S, Wang Y et al (2009) Autophagy regulates lipid metabolism. Nature 458:1131–1135
Stromhaug PE, Klionsky DJ (2001) Approaching the molecular mechanism of autophagy. Traffic 2:524–531
Glick D, Barth S, Macleod KF (2010) Autophagy: cellular and molecular mechanisms. J Pathol 221:3–12
Singh R, Cuervo Ana M (2011) Autophagy in the cellular energetic balance. Cell Metab 13:495–504
Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel Yves L (2011) Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab 13:655–667
Cahova M, Dankova H, Palenickova E, Papackova Z, Kazdova L (2010) The autophagy-lysosomal pathway is involved in TAG degradation in the liver: the effect of high-sucrose and high-fat diet. Folia Biol 56:173–182
Cahova M, Dankova H, Palenickova E et al (2012) The increased activity of liver lysosomal lipase in nonalcoholic fatty liver disease contributes to the development of hepatic insulin resistance. Biochem Res Int 2012:11
Miyazaki J-I, Araki K, Yamato E et al (1990) Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127:126–132
Busch AK, Cordery D, Denyer GS, Biden TJ (2002) Expression profiling of palmitate- and oleate-regulated genes provides novel insights into the effects of chronic lipid exposure on pancreatic beta-cell function. Diabetes 51:977–987
Boslem E, MacIntosh G, Preston AM et al (2011) A lipidomic screen of palmitate-treated MIN6 β-cells links sphingolipid metabolites with endoplasmic reticulum (ER) stress and impaired protein trafficking. Biochem J 435:267–276
Vázquez CL, Colombo MI (2009) Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. In: Daniel JK (ed) Methods in enzymology. Academic, Waltham, pp 85–95
Meikle PJ, Wong G, Tsorotes D et al (2011) Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler Thromb Vasc Biol 31:2723–2732
Rosenbaum AI, Cosner CC, Mariani CJ, Maxfield FR, Wiest O, Helquist P (2010) Thiadiazole carbamates: potent inhibitors of lysosomal acid lipase and potential Niemann–Pick type C disease therapeutics. J Med Chem 53:5281–5289
Rosenbaum AI, Rujoi M, Huang AY, Du H, Grabowski GA, Maxfield FR (2009) Chemical screen to reduce sterol accumulation in Niemann–Pick C disease cells identifies novel lysosomal acid lipase inhibitors. Biochim Biophys Acta 1791:1155–1165
Cnop M, Hannaert JC, Hoorens A, Eizirik DL, Pipeleers DG (2001) Inverse relationship between cytotoxicity of free fatty acids in pancreatic islet cells and cellular triglyceride accumulation. Diabetes 50:1771–1777
Busch AK, Gurisik E, Cordery DV et al (2005) Increased fatty acid desaturation and enhanced expression of stearoyl coenzyme A desaturase protects pancreatic beta-cells from lipoapoptosis. Diabetes 54:2917–2924
Larsson S, Resja S, Gomez MF, James P, Holm C (2012) Characterization of the lipid droplet proteome of a clonal insulin-producing β-cell line (INS-1 832/13). J Proteome Res 11:1264–1273
Kaushik S, Rodriguez-Navarro Jose A, Arias E et al (2011) Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab 14:173–183
Kovsan J, Bashan N, Greenberg AS, Rudich A (2009) Potential role of autophagy in modulation of lipid metabolism. Am J Physiol Endocrinol Metab 298:E1–E7
Sieber MH, Thummel CS (2012) Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the Drosophila LipA homolog magro. Cell Metab 15:122–127
Nolan CJ, Madiraju MS, Delghingaro-Augusto V, Peyot ML, Prentki M (2006) Fatty acid signaling in the beta-cell and insulin secretion. Diabetes 55(suppl 2):S16–S23
Peyot M-L, Nolan CJ, Soni K et al (2004) Hormone-sensitive lipase has a role in lipid signaling for insulin secretion but is nonessential for the incretin action of glucagon-like peptide 1. Diabetes 53:1733–1742
O’Rourke EJ, Ruvkun G (2013) MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 16:668–676
Settembre C, de Cegli R, Mansueto G et al (2013) TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 16:647–658
Herrero L, Rubi B, Sebastian D et al (2005) Alteration of the malonyl-CoA/carnitine palmitoyltransferase I interaction in the beta-cell impairs glucose-induced insulin secretion. Diabetes 54:462–471
Kwan EP, Xie L, Sheu L et al (2006) Munc13-1 deficiency reduces insulin secretion and causes abnormal glucose tolerance. Diabetes 55:1421–1429
Schmitz-Peiffer C, Biden TJ (2008) Protein kinase C function in muscle, liver, and beta-cells and its therapeutic implications for type 2 diabetes. Diabetes 57:1774–1783
Ebato C, Uchida T, Arakawa M et al (2008) Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 8:325–332
Jung HS, Chung KW, Won Kim J et al (2008) Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab 8:318–324
Tanemura M, Ohmura Y, Deguchi T et al (2012) Rapamycin causes upregulation of autophagy and impairs islets function both in vitro and in vivo. Am J Transplant 12:102–114
Acknowledgements
We thank M. Cahova (Institute for Clinical and Experimental Medicine, Prague, Czech Republic) for suggesting the use of Lalistat, and L. Marshall (Garvan Institute of Medical Research, Darlinghurst, NSW, Australia) for help in setting up the Nile Red staining protocol. We are grateful to L. O’Reilly and R. Laybutt (Garvan Institute of Medical Research, Darlinghurst, NSW, Australia) for islet isolation and for critical review of the manuscript, respectively.
Funding
This work was supported by a project grant (TJB), research fellowships (TJB and PJM) and a post-graduate scholarship (GLP) from the National Medical Research Council of Australia.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
GLP, NM, KYC, PJM, JC and AD performed experiments, analysed data, contributed to the acquisition of data, and to the revision of the manuscript. CCC, PB and PH contributed to the acquisition of data, and the revision of the manuscript. TJB and PJM contributed to the revision of the manuscript. GLP wrote the manuscript and designed experiments. TJB conceived and designed experiments and wrote the manuscript. All authors approved the final version of this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pearson, G.L., Mellett, N., Chu, K.Y. et al. Lysosomal acid lipase and lipophagy are constitutive negative regulators of glucose-stimulated insulin secretion from pancreatic beta cells. Diabetologia 57, 129–139 (2014). https://doi.org/10.1007/s00125-013-3083-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-013-3083-x