Abstract
Aims/hypothesis
ALR/Lt, a mouse strain with strong resistance to type 1 diabetes, is closely related to autoimmune type 1 diabetes-prone NOD/Lt mice. ALR pancreatic beta cells are resistant to the beta cell toxin alloxan, combinations of cytotoxic cytokines, and diabetogenic NOD T-cell lines. Reciprocal F1 hybrids between either ALR and NOD or ALR and NON/Lt, showed that alloxan resistance was transmitted to F1 progeny only when ALR was the maternal parent. Here we show that the mitochondrial genome (mtDNA) of ALR mice contributes resistance to diabetes.
Methods
When F1 progeny from reciprocal outcrosses between ALR and NOD were backcrossed to NOD, a four-fold lower frequency of spontaneous type 1 diabetes development occurred when ALR contributed the mtDNA. Because of the apparent interaction between nuclear and mtDNA, the mitochondrial genomes were sequenced.
Results
An ALR-specific sequence variation in the mt-Nd2 gene producing a leucine to methionine substitution at amino acid residue 276 in the NADH dehydrogenase 2 was discovered. An isoleucine to valine mutation in the mt-Co3 gene encoding COX3 distinguished ALR and NOD from NON and ALS. All four strains were distinguished by variation in a mt-encoded arginyl tRNA polyadenine tract. Shared alleles of mt-Co3 and mt-Tr comparing NOD and ALR allowed for exclusion of these two genes as candidates, implicating the mt-Nd2 variation as a potential ALR-derived type 1 diabetes protective gene.
Conclusions/interpretation
The unusual resistance of ALR mice to both ROS-mediated and autoimmune type 1 diabete stresses reflects an interaction between the nuclear and mt genomes. The latter contribution is most likely via a single nucleotide polymorphism in mt-Nd2.
Similar content being viewed by others
Introduction
Mitochondria (mt) play key roles in cellular energy production and cell death, as well as representing the major source of intracellularly generated oxygen free radicals during normal metabolism [1]. Peroxidation by reactive oxygen species (ROS) of mt cardiolipin leads to dissociation of cytochrome c from the mt inner membrane, a step required for initiating apoptotic cell death [2]. As intracellular concentrations of ROS rise, nuclear genes encoding antioxidant defenses, including the phospholipid hydroperoxide glutathione peroxidase (Gpx4), classical glutathione peroxidase (Gpx) and manganese superoxide dismutase (Sod2) are expressed [2]. Hence, an important axis exists between the nuclear and mitochondrial genomes in maintaining intracellular redox potential. Insulin biosynthetic and secretory capacities of pancreatic beta cells are highly dependent upon normal nuclear/mt communication [3], mt ATP generation [4], and exquisite sensitivity to ROS-mediated damage [5, 6]. In this context, reports of mtDNA mutations associated with matrilineal type 2 diabetes (T2D) pedigrees in humans and rodents are becoming increasingly more frequent [7–9], accounting for up to 1% of human T2D. The mtDNA mutations are heteroplasmic in all of the described cases, and most commonly involve the A3243G mutation in the gene encoding mt tRNALeuUUR (mt-Tl1) [7]. A T14577C missense mutation in the mt-Nd6 gene encoding the complex I NADH dehydrogenase subunit 6 has also been identified as a pathogenic mutation associated with maternally inherited T2D in a Japanese population [10]. Cumulative mutations in mtDNA presumably contribute to the decline in mt function accompanying progressive development of insulin resistance associated with human aging [11]. Mutations in mtDNA are not commonly associated with susceptibility or resistance to autoimmune type 1 diabetes (T1D) in humans, although a recently reported C5178A transversion resulting in a leucine to methonine substitution in the mt mt-Nd2 gene has been associated with protection from T1D in an autoantibody-positive Japanese cohort [12].
Given the importance of maintaining normal redox potential in beta cells, it is not surprising that experimental overexpression of free radical dissipating enzymes such as catalase, glutathione peroxidase and the cytoplasmic copper–zinc superoxide dismutase protects cultured rodent beta cells from deleterious combinations of cytokines (interferon gamma [IFN-γ], tumor necrosis factor α [TNF-α] and interleukin 1 [IL-1]) [13]. However, it is unusual to find a mouse with systemically upregulated antioxidant defences that include increased expression at the beta cell level as well. The ALR/Lt mouse, originally selected for resistance to alloxan, a beta cell selective toxin that generates hydroxyl radicals [14], provides a rare example. We demonstrated that the molecular basis for this resistance was a systemic upregulation of molecules and enzymes associated with free radical dissipation in unmanipulated ALR/Lt mice [15, 16]. ALR/Lt males exhibited elevated ratios of reduced glutathione to oxidized glutathione (GSH/GSSG) in the blood and plasma when compared to untreated ALS/Lt mice (the latter coselected in Japan for high sensitivity to alloxan-induced diabetes). The lower oxidative burden in ALR/Lt mice correlated with significantly higher specific activities for a battery of antioxidant enzymes not only in comparison to ALS mice, but also in comparison to the closely related NOD/Lt strain that develops spontaneous T-cell-mediated T1D [16, 17]. ALR pancreatic beta cells, both in vivo and in vitro, were resistant not only to alloxan, but also resistant in vitro to the cytotoxic combination of IFN-γ, TNF-α and IL-1. Strikingly, although expressing the appropriate MHC class I restriction element required for recognition by diabetogenic NOD CD8+ T cells, ALR/Lt beta cells were resistant to destruction by such diabetogenic T cells both in vitro and in vivo [17].
Outcross of ALR/Lt with both ALS/Lt and NOD/Lt demonstrated that F1 hybrid protection from alloxan-induced diabetes in the case of ALS/Lt outcross, and spontaneous development of T1D in the case of NOD/Lt outcross, was transmitted as a dominant trait [16, 17]. A 1:1 segregation of alloxan-sensitive versus alloxan-resistant males was observed following backcross of alloxan-resistant (ALS×ALR)F1 females to the alloxan-sensitive ALS males [16]. However, we were unable to map the gene or genes controlling alloxan sensitivity in this cross because of the subsequently discovered complication that ALS/Lt males spontaneously developed glucose intolerance, insulin resistance and T2D [18]. In this first backcross (BC1) progeny, a unique ALR allele on chromosome 3 (Chr. 3) was mapped that suppressed neutrophil-generated superoxide burst following activation [19]. In outcross of NOD females with ALR males, and backcross of the T1D-resistant females to NOD males, three nuclear-encoded genes from the ALR genome were mapped that suppressed T1D development in BC1 females [20]. One resistance locus colocalized to the above-mentioned Chr. 3 region containing the suppressor of superoxide production (Susp) allele; one was MHC-linked on Chr. 17, and one mapped on Chr. 8 [20]. Because F1 mice from reciprocal ALR and ALS crosses were resistant to alloxan-induced diabetes, and F1 mice from reciprocal crosses of ALR and NOD mice were resistant to spontaneous T1D development, protective contributions from the ALR mitochondrial genome were not immediately indicated.
Recently, we identified a pathogenic mtDNA mutation in strain A/J mice that interacted with a nuclear gene on Chr. 10, designated Ahl, that produced a maternally inherited, age-related hearing loss [21]. An adenine insertion in a polyadenine repeat in the mitochondrial arginyl tRNA (mt-Tr) gene of the A/J mt genome was associated with this strain’s hearing impairment. The mt tRNA-Arg association was not demonstrated in outcrosses between CAST/Ei and NOD [21], yet NOD, ALR, and ALS, but not NON, all exhibit the age-related hearing loss phenotype [22]. In addition to sharing the pathogenic Ahl1 allele with A/J on Chr. 10, NOD was found to segregate an additional nuclear gene on Chr. 5 (Ahl2) [23, 24]. These findings raised the possibility that strain-specific mutations in the ALR mt genome might differentiate this strain from the three related strains. If so, given the important role of mt in the generation of endogenous ROS, such a difference might represent a component of the unusual diabetes resistance of ALR/Lt mice. Since we had previously completed a segregation analysis of ALR/NOD nuclear genes segregating for type 1 diabetes susceptibility/resistance when the mt donor was the NOD/LtDvs female, the current study was undertaken to determine whether the ALR mt genome could contribute to type 1 diabetes resistance.
Materials and methods
Mice
ALR/Lt, ALS/Lt, NOD/Lt, NOD/LtDvs and NON/Lt mice used in this study were bred and maintained in the animal research facility at the Jackson Laboratory, Bar Harbor, ME, USA, or the Rangos Research Center animal facility, Pittsburgh, PA, USA. NOD/LtJ and C57BL/6J were obtained from Jax Mice and Services (Jackson Laboratory, Bar Harbor, ME, USA) and housed in the Rangos Research Center animal facility. NOD/LtDvs, a recent substrain derived from NOD/Lt at the Jackson Laboratory, has a type 1 diabetes frequency in mice of both sexes indistinguishable from that in NOD/Lt or the commercially available NOD/LtJ. All F1 mice generated by outcrosses with NOD/Lt or NOD/LtDvs were free of spontaneously developing autoimmune diabetes. The mice were bred and maintained in a specific pathogen-free vivarium and allowed free access to food (autoclaved diet NIH-31, 6% fat, PMI, St Louis, MO, USA) and acidified drinking water. Direction of crosses is indicated in the tables, with the maternal parent indicated first. All procedures involving the use of animals were approved by the animal care and use committee of either the Jackson Laboratory or the Children’s Hospital of Pittsburgh and were in compliance with the “Principles of Laboratory Animal Care” and the current laws of the United States.
mtDNA sequencing
Nuclear and mitochondrial DNA were co-purified from kidney and liver of ALR/Lt, ALS/Lt, NOD/Lt, NOD/LtDvs, NOD/LtJ, NON/Lt, C57BL/6J, as well as F1 and BC1 mice by standard phenol–chloroform extraction methodology as previously described [25]. Synthetic oligonucleotides were designed using the Oligo 6.3 program (Molecular Biology Insights, Cascade, CO, USA) to amplify the whole mtDNA in 21 overlapping PCR products with an average size of 914 bp. PCR reactions contained 400 ng DNA, 50 pmol of each primer, 10 mM Tris–HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, and 1 U Taq DNA Polymerase (Applied Biosystems, Foster City, CA, USA) in a total volume of 50 μl. The DNA was initially denatured at 94°C for 5 min, followed by 35 step cycles of denaturing at 94°C for 30 s, annealing at 49–53°C (according to the conditions for each primer pair recommended by the Oligo program) for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min in the GeneAmp PCR System 9700 (Applied Biosystems). PCR products were extracted after electrophoresis on 1% agarose gel using the Concert Rapid Gel Extraction System (Invitrogen, Carlsbad, CA, USA) or directly purified using the Concert Rapid PCR Purification System (Invitrogen).
Complete sequence was obtained by sequencing each of the PCR products in both directions with primers previously used for the PCR amplifications. The dideoxynucleotide chain termination method was employed by using dsDNA Cycle Sequencing System (Invitrogen) with γ-[33P]-dATP (NEN). Some sequencing reactions were performed using BD v3.1 kits (Applied Biosystems) and a PRISM 3100 Genetic Analyzer (Applied Biosystems).
Genetic control of alloxan (AL)-induced diabetes
Seven-week-old ALR/Lt, ALS/Lt, NOD/Lt, NON/Lt males, reciprocal F1 males from outcrosses of the ALR strain with either ALS/Lt, NOD/Lt or NON/Lt strains, as well as BC1 males, were weighed and a blood sample taken retro-orbitally before AL treatment and at 7 days postinjection. Alloxan monohydrate (Sigma, St Louis, MO, USA) was diluted in sterile PBS immediately prior to i.v. administration and given at a concentration of 52 mg/kg. Controls received PBS only. Capillary tubes containing blood were centrifuged in a Micro-MB Centrifuge (Thermo IEC, Needham Heights, MA, USA) for 2 min, and plasma was frozen until assayed. Plasma glucose values were determined using a Beckman Glucose Analyzer II (Beckman Coulter, Fullerton, CA, USA).
Development of spontaneous type 1 diabetes in ALR outcrosses to NOD
Diabetes development was monitored by biweekly testing for glucosuria in BC1 females from 8 until 40 weeks of age (Diastix; a kind gift of Bayer, Elkhart, IN, USA). The 120 female progeny produced in the (NOD/LtDvs×ALR/Lt)F1×NOD outcross are the same mice reported previously [20]; the diabetes frequency for this cross is reproduced here only for comparison to the diabetes frequency for 107 comparably aged [(ALR/Lt×NOD/Lt)F1×NOD/Lt] BC1 females generated from the reciprocal cross in the present study. Clinical diabetes was diagnosed after two sequential positive tests. Diabetic females were euthanized by CO2 asphyxiation. Non-glycosuric females were euthanized at the 40-week age point, and pancreata were removed and assessed for extent of insulitic damage to islets using a scoring system described elsewhere [20]. Segregants with clinical diabetes of at least 2-week duration typically exhibited mean insulitis scores of 3.5–4.0 (out of a maximally severe score of 4.0). Hence, segregants that were non-glycosuric at the time of euthanasia, but whose pancreata presented mean insulitis scores ≥3.8 were classified as incipient diabetics [20].
SNP analysis
DNA was prepared from kidney by standard phenol–chloroform extraction methodology as above. SNP detection was performed using pyrosequencing. To evaluate the SNPs in mt-Nd2 and mt-Tr, genotyping was performed on all BC1 mice plus controls by PCR amplification of the region surrounding these SNP. We did not develop a test for the SNP at position 9,348, as ALR and NOD have identical alleles of mt-Co3. The initial PCR amplified either a 395-bp product for mt-Nd2 (mt-Nd2-F 5′-TCA ACT CAA TCT CAC TTC TAT G and mt-Nd2-R-Biotin 5′-Biotin-GCT TTG AAG GCT CGC GGA C) or 522-bp product for mt-Tr (mt-Tr-F 5′-GAC CCT ACA AGC TCT GCA C and mt-Tr-R-Biotin 5′-Biotin-AGT CCT ACA GCT GCT TCG). The DNA was denatured at 94°C for 5 min, followed by 45 step cycles of denaturing at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min in the GeneAmp PCR System 9700 (Applied Biosystems). Pyrosequencing was performed as previously described [26] using mt-Nd2-PSQ 5′-TTATAAAAAACAACTGT and a dispensation order of G-A-C-T-A-T-A-T-A-G-C. For mt-Tr a trio of primers (mt-Tr-T0 [GAA ATC ATT AAT TTT TTT T], mt-Tr-T1 [GAA ATC ATT AAT TTT TTT TT], and mt-Tr-T2 [GAA ATC ATT AAT TTT TTT TTT]) were used with a dispensation order G-T-A-C-T-A-T-A-C-A-T. Data was collected and analyzed used a PSQ96 (Biotage AB and Biosystems/Pyrosequencing, Uppsala, Sweden).
Statistical analyses
All values reported are means ± SD calculated with an n of at least 6. Significance was determined with a one-way ANOVA using SuperANOVA for the Macintosh (Abacus Concepts, Berkeley, CA, USA). Chi-square was calculated using StatView 5.0.1 (SAS Institute, Cary, NC, USA). Differences in plasma glucose values and the genetic analysis were considered significant at p<0.05 and p<0.0005, respectively.
Results
Alloxan-induced diabetes
Data in Table 1 show, as expected, that among males of the four related parental strains (ALR, ALS, NON, and NOD) used to generate F1 hybrids, only the ALR/Lt males were resistant to AL-induced hyperglycemia. Similarly, among females, only ALR/Lt females were AL-resistant (data not shown). As observed previously, the ALR nuclear genome conferred dominant resistance in outcrosses with ALS/Lt mice regardless of whether ALR/Lt contributed the maternal or the paternal genome (Table 1) [16]. In contrast, when ALR/Lt mice were mated with either NOD/Lt or NON/Lt mice, AL resistance required that ALR/Lt be the maternal parent.
Spontaneous type 1 diabetes incidence in reciprocal backcross populations
Comparative frequencies of spontaneous type 1 diabetes development in females following backcross of [NOD/LtDvs (♀)×ALR/Lt (♂)]F1 females with NOD/LtDvs males, and reciprocally [ALR/Lt (♀)×NOD/Lt (♂)]F1 females with NOD/Lt males supported the hypothesis that the ALR mt genome (rather than genetic imprinting), in combination with ALR nuclear-encoded genes previously demonstrated by linkage analysis [20], contributed to the unusual degree of resistance to type 1 diabetes inherent to this strain. As shown in Table 2, a four-fold higher frequency of diabetes (both clinically manifested glycosuria and incipient diabetes based upon extremely high mean insulitis score) developed when the mt genome derived from the NOD maternal lineage as compared to the ALR maternal lineage.
mtDNA sequencing
Mitochondrial genome scans were performed to identify polymorphisms distinguishing the mt populations in all four related strains (Table 3). Full mitochondrial chromosome sequences were submitted to GenBank and have been given the following accession numbers: ALR/Lt [AY533105], ALS/Lt [AY533106], NOD/Lt and NOD/LtDvs [AY533107], and NON [AY533108]. The ALR/Lt mt genome was distinguished from all other related strains by a transversion in the mt-Nd2 gene [A4738C] producing a non-conservative M to L substitution at amino acid residue 276 in the NADH dehydrogenase-2 subunit. The ALR/Lt and both the NOD/Lt and NOD/LtDvs substrains differed from both the NON and ALS strains by a conservative A9827G transversion producing an I to V change at amino acid residue 248 in the mt-Co3 gene encoding COX3. All four strains were distinguished by variation in the mt-Tr polyA tract previously associated with age-associated deafness in A/J mice.
SNP detection in BC1 populations
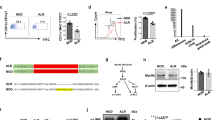
To determine the extent to which mitochondrial genotypes were maternally transmitted as well as the presence of homoplasmy, we developed assays for the SNPs at nt 4,738 and 9,827. During validation of both pyrosequencing assays, the parental strains were surveyed for potential heteroplasmy. For both SNPs, we evaluated at least six mice per strain. Testing for the A4738C SNP in the mt-Nd2 gene distinguishing the ALR mt genome from that of the other related strains established that all parental strains were homoplasmic, with all strains except ALR/Lt encoding a “C” at this position (Table 3 and Fig. 1). As expected, BC1 mice encoded the allele in a homoplasmic state that was transmitted through the maternal lineage (Fig. 1), with all [(NOD/LtDvs (♀)×ALR/Lt (♂)) F1 (♀)×NOD/LtDvs (♂)] BC1 coding for a C and all [(ALR/Lt (♀)×NOD/Lt (♂)) F1 (♀)×NOD/Lt (♂)] maintaining an A at nt 4,738. Figure 1 also demonstrates that mtDNA was inherited through the maternal lineage. Diabetes was significantly skewed to BC1 mice with the NOD or “C” allele (32/120) compared to the ALR or “A” allele (7/107). Linkage analysis calculated a chi-square of 16.1 (p<0.0001).
SNP typing for the mt-Nd2 allele resulted in distinct pyrosequencing pyrograms for the two alleles. Pyrosequencing was performed using a sequencing primer that annealed over positions 4,721–4,737, utilizing the SNP at position 4,738 as the first nucleotide to be incorporated. This strategy created an out-of-phase sequencing reaction, allowing for distinct pyrograms for each allele. The pyrograms distinguished between the two alleles encoded by a ALR (mt-ND2a-ATAATTATAGC), and b NOD (mt-Nd2b-CTAATTATAGC). The polymorphic residue is indicated by an underlined A or C. Dashes represent nucleotide dispensations that were not incorporated into the nascent chain. Pyrosequencing allowed for determination of the inheritance of the “a” allele in (c) ([ALR×NOD]×NOD) BC1 and the “b” allele in (d) ([NOD×ALR]×NOD) BC1.
With regard to the mt-Tr length polymorphism in the polyA tract, we did not detect heteroplasmy in any of the ALR/Lt (9A), NON/Lt, (8A) ALS/Lt (10A), NOD/LtDvs (9A), or NOD/LtJ (10A) samples tested. While we did not detect heteroplasmy in the NOD/Lt strain, the strain from which both the NOD/LtDvs and NOD/LtJ were derived, we did detect the presence of both the 9A and 10A alleles in homoplasmy. Of the 12 NOD/Lt mice we examined, the mt genomes from ten encoded the 9A allele while the mt genomes of the other two encoded the 10A allele. We also determined that all of the reciprocal BC1 mice (both [(NOD/LtDvs (♀)×ALR/Lt (♂)) F1 (♀)×NOD/LtDvs (♂)] and [(ALR/Lt (♀)×NOD/Lt (♂)) F1 (♀)×NOD/Lt (♂)]) were homoplasmic for the 9A allele. Hence, all parental progenitors in both crosses were homoplasmic for the mt-Tr 9A variant.
Discussion
That the mt population from ALR/Lt mice is unusual in terms of its resistance to oxidative stress has recently been confirmed by Turko et al. [27] who analyzed nitration of heart mt proteins from ALR/Lt and ALS/Lt mice following AL treatment in vivo, or after incubation of purified mt from untreated mice in vitro with either peroxynitrite or a combination of sodium nitrite/H2O2/myeloperoxidase. Whereas both soluble and membrane localized proteins of ALS/Lt mt, including succinyl-CoA:3-oxoacid CoA transferase and the NADH-ubiquinone oxidoreductase (complex I) were tyrosine nitrosylated, the same mt proteins from ALR/Lt were not nitrosylated [26]. The previous mapping of the unique ALR/Lt suppressor of superoxide production (Susp) allele to Chr. 3 [19] in the same interval that segregated for resistance to type 1 diabetes in the [(NOD/LtDvs (♀)×ALR/Lt (♂))×NOD/LtDvs (♂)] BC1 segregation analysis [20] suggests the contributions of nuclear genes to the remarkable antioxidant defense system of ALR/Lt mice. However, a major contribution from the ALR/Lt mt genome was manifest by a profound reduction in type 1 diabetes frequency when the ALR/Lt rather than the NOD/Lt female was the source of the mt DNA in outcross/backcross analysis (Table 2). Indeed, the diabetes-suppressive effect of the ALR/Lt mt genome was so pronounced that mapping of nuclear-encoded resistance alleles was precluded because too few diabetic probands (7/107 females) were recovered.
The molecular basis for the upregulated antioxidant defenses of ALR/Lt mice remains to be elucidated. Data in Table 3 demonstrating multiple polymorphisms distinguishing the ALR/Lt mt genome from that of NOD, NON, and ALS implicate an ALR-specific interaction of nuclear and mt genomes. Nuclear genes encoding key elements of islet antioxidant defenses, such as the heme oxygenase 1 gene, a candidate gene for the ALR-contributed resistance locus on Chr. 8 [20], are induced by reactive oxygen species generated by the mt electron transport chain [28]. An interesting candidate gene for Susp (and the ALR-contributed type 1 diabetes resistance locus on Chr. 3) is Etfdh, a nuclear gene encoding the electron transport flavoprotein-ubiquinone dehydrogenase. This enzyme transfers reducing equivalents from fatty acid oxidation in the mt matrix to the electron transport chain. Reduced activity of this enzyme in ALR/Lt cells could conceivably produce a higher electron leak into the cytoplasm, thereby activating nuclear gene transcription of multiple products contributing to elevated antioxidant defense. In this regard, the apparently unique non-conservative replacement of leucine by methionine at amino acid residue 276 in the ALR/Lt mt-Nd2 (Table 3) via an A to C nucleotide substitution (Fig. 1), may well represent a component of the mt-derived resistance. This inference is reinforced by the association of human mt-Nd2 mutations not only with T2D in Japanese, but also with type 1 diabetes [12]. In an islet cell antibody positive Japanese population, a mutation in the mt-Nd2 gene changing the L at amino acid residue 237 to a M correlated with protection from type 1 diabetes [12]. The polymorphism in the mt genome-encoded Cox3 subunit (Table 3) is an unlikely contributor, as the resistant ALR genome shares this substitution with the NOD genome. The significance of the polymorphisms in the mt-Tr gene (Table 3) is presently unclear; this mt gene was not demonstrated to be interactive with nuclear genes (Ahl1, Ahl2) contributing to hearing loss in a NOD×CAST cross [21, 24]. Interestingly, however, the normally hearing NON/Lt strain shares the 8-bp polyA polymorphism with the normally hearing CAST/Ei strain. Despite an A/J strain allele-specific interaction with the nuclear genome to effect age-related hearing loss, only nuclear genes have thus far been linked to the hearing loss of NOD/Lt mice, a phenotype shared with ALR/Lt and ALS/Lt. Further, NOD substrains differ in the allele they maintain, with NOD/LtDvs encoding the 9A allele, NOD/LtJ the 10A allele, and mice of the NOD/Lt encoding either the 9A allele or 10A allele in homoplasmy. In fact all mice tested were homoplasmic over the mtDNA, which differs from a previous report of heteroplasmy in mt-Tr of NOD/Lt mice [21]. Type 1 diabetes frequency is not different when comparing the NOD/Lt, NOD/Dvs, and NOD/LtJ (data not shown). Therefore, we infer that homoplasmic variation in the polyA tract of mt-Tr differentiating these three NOD substrains is not a critical determinant of type 1 diabetes susceptibility.
To better understand the role of the mt-Nd2 SNP in diabetes protection, we have begun to generate reciprocal lines of mtDNA consomic mice. These lines will possess the nuclear genome of either ALR and the mtDNA from NOD (ALR.NODmt) or the inverse (NOD.ALRmt). Crosses of the NOD.ALRmt with NOD mice that are congenic for ALR-derived type 1 diabetes-protective loci [20] will enable us to perform experiments detailing the nuclear–mitochondrial interactions that are important in diabetes protection.
In summary, we have provided evidence that one component of the markedly elevated defense of ALR/Lt mice against free radical mediated stresses most likely entails a specific mutation in mt-Nd2. These findings of a nuclear/mitochondrial genomic interaction add to our understanding of the complex genetic basis of spontaneous type 1 diabetes development in NOD mice [29].
Abbreviations
- ALR:
-
Alloxan resistant
- ALS:
-
Alloxan susceptible
- BC1:
-
Backcross 1
- mt-Co3:
-
Mitochondrial cytochrome c oxidase III
- mt-Nd2:
-
Mitochondrial NADH dehydrogenase 2
- NOD:
-
Nonobese diabetic
- NON:
-
Nonobese nondiabetic
References
Genova ML, Pich MM, Biondi A et al (2003) Mitochondrial production of oxygen radical species and the role of coenzyme Q as an antioxidant. Exp Biol Med 228:506–513
Nomura K, Imai H, Koumura T, Nakagawa Y (2001) Involvement of mitochondrial phospholipid hydroperoxide glutathione peroxidase as an antiapoptotic factor. Biol Signals Recept 10:81–92
Silva JP, Kohler M, Graff C et al (2000) Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet 26:336–340
Wollheim CB, Maechler P (2002) β-Cell mitochondria and insulin secretion: messenger role of nucleotides and metabolites. Diabetes 51[Suppl 1]:S37–S42
Nishikawa T, Edelstein D, Du XL et al (2000) Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature 404:787–790
Robertson RP, Harmon J, Tran PO, Tanaka Y, Takahashi H (2003) Glucose toxicity in beta-cells: type 2 diabetes, good radicals gone bad, and the glutathione connection. Diabetes 52:581–587
Maassen J, Vandenou J, Thart L, Lemkes H (1997) Maternally inherited diabetes and deafness—a diabetic subtype associated with a mutation in mitochondrial-DNA. Horm Metab 29:50–55
Mathews CE, Berdanier CD (1998) Diabetes mellitus, a mitochondrial genetic disease. PSEBM 219:97–108
Fischel-Ghodsian N (2001) Mitochondrial DNA mutations and diabetes: another step toward individualized medicine. Ann Intern Med 134:777–779
Tawata M, Hayashi JI, Isobe K et al (2000) A new mitochondrial DNA mutation at 14577 T/C is probably a major pathogenic mutation for maternally inherited type 2 diabetes. Diabetes 49:1269–1272
Petersen KF, Befroy D, Dufour S et al (2003) Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300:1140–1142
Uchigata Y, Okada T, Gong JS, Yamada Y, Iwamoto Y, Tanaka M (2002) A mitochondrial genotype associated with the development of autoimmune-related type 1 diabetes. Diabetes Care 25:2106
Azevedo-Martins AK, Lortz S, Lenzen S, Curi R, Eizirik DL, Tiedge M (2003) Improvement of the mitochondrial antioxidant defense status prevents cytokine-induced nuclear factor-kappaB activation in insulin-producing cells. Diabetes 52:93–101
Sekiguchi F, Ishibashi K, Katoh H, Kawamoto Y, Ino T (1990) Genetic profile of alloxan-induced diabetes-susceptible mice (ALS) and resistant mice (ALR). Exp Anim 39:269–272
Mathews CE, Leiter EH (1999) Constitutive differences in anti-oxidant defense status distinguishes alloxan resistant (ALR/Lt) and alloxan susceptible (ALS/Lt) mice. Free Radical Biol Med 27:449–455
Mathews CE, Leiter EH (1999) Resistance of ALR/Lt islets to free radical-mediated diabetogenic stress is inherited as a dominant trait. Diabetes 48:2189–2196
Mathews CE, Graser R, Savinov A, Serreze DV, Leiter EH (2001) The NOD/Lt-related ALR/Lt strain: unusual resistance of beta cells to autoimmune killing uncovers a role for beta-cell expressed resistance determinants. Proc Natl Acad Sci U S A 98:235–240
Mathews CE, Leiter EH (2003) ALS/Lt: A new Type 2 diabetes mouse model associated with low free radical scavenging potential. Diabetes 52:S125–S129
Mathews CE, Dunn BD, Hannigan MO, Huang CK, Leiter EH (2002) Genetic control of neutrophil superoxide production in diabetes-resistant ALR/Lt mice. Free Radic Biol Med 32:744–751
Mathews CE, Graser RT, Bagley RJ et al (2003) Genetic analysis of resistance to type 1 diabetes in ALR/Lt mice, a NOD-related strain with defenses against autoimmune-mediated diabetogenic stress. Immunogenetics 55:491–496
Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N (2001) A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nat Genet 27:191–194
Zheng QY, Johnson KR, Erway LC (1999) Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res 130:94–107
Johnson KR, Zheng QY, Erway LC (2000) A major gene affecting age-related hearing loss is common to at least ten inbred strains of mice. Genomics 70:171–180
Johnson KR, Zheng QY (2002) Ahl2, a second locus affecting age-related hearing loss in mice. Genomics 80:461–464
Mathews CE, McGraw RA, Dean R, Berdanier CD (1999) Inheritance of a mitochondrial DNA defect and impaired glucose tolerance in BHE/Cdb rats. Diabetelogia 42:35–40
Ringquist S, Alexander AM, Styche A, Pecoraro C, Rudert WA, Trucco M (2004) HLA class II DRB high resolution genotyping by pyrosequencing: comparison of group specific PCR and pyrosequencing primers. Hum Immunol 65:163–174
Turko IV, Li L, Aulak KS, Stuehr DJ, Chang JY, Murad F (2003) Protein tyrosine nitration in the mitochondria from diabetic mouse heart: implications to dysfunctional mitochondria in diabetes. J Biol Chem 278:33972–33977
Chang SH, Garcia J, Melendez JA, Kilberg MS, Agarwal A (2003) Heme oxygenase 1 gene induction by glucose deprivation is mediated by reactive oxygen species via the mitochondrial electron-transport chain. Biochem J 371:877–885
Serreze DV, Leiter EH (2001) Genes and pathways underlying autoimmune diabetes in NOD mice. In: von Herrath M (ed) Molecular pathology of insulin-dependent diabetes mellitus. Karger, New York, pp 31–67
Acknowledgements
We gratefully acknowledge support of the Juvenile Diabetes Foundation International, the American Diabetes Association, and National Institutes of Health grants F32DK09865 (C.E.M.), AI056374 (C.E.M.), DK27722 (E.H.L.), DK36175 (E.H.L.), DC01402 (N.F.-G.), and DC04092 (N.F.-G.). TJL Institutional shared services were supported by National Cancer Institute Center Support Grant CA-34196. We thank Mr Steve Langley, Ms Rebecca Bagley, Ms Darcy Pomerleau, Ms Therese Leibert, Ms Alex Styce, and Dr Massimo Trucco for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mathews, C.E., Leiter, E.H., Spirina, O. et al. mt-Nd2 Allele of the ALR/Lt mouse confers resistance against both chemically induced and autoimmune diabetes. Diabetologia 48, 261–267 (2005). https://doi.org/10.1007/s00125-004-1644-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-004-1644-8