Abstract
Key message
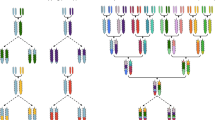
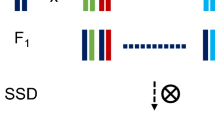
MAGIC populations present novel challenges and opportunities in crops due to their complex pedigree structure. They offer great potential both for dissecting genomic structure and for improving breeding populations.
Abstract
The past decade has seen the rise of multiparental populations as a study design offering great advantages for genetic studies in plants. The genetic diversity of multiple parents, recombined over several generations, generates a genetic resource population with large phenotypic diversity suitable for high-resolution trait mapping. While there are many variations on the general design, this review focuses on populations where the parents have all been inter-mated, typically termed Multi-parent Advanced Generation Intercrosses (MAGIC). Such populations have already been created in model animals and plants, and are emerging in many crop species. However, there has been little consideration of the full range of factors which create novel challenges for design and analysis in these populations. We will present brief descriptions of large MAGIC crop studies currently in progress to motivate discussion of population construction, efficient experimental design, and genetic analysis in these populations. In addition, we will highlight some recent achievements and discuss the opportunities and advantages to exploit the unique structure of these resources post-QTL analysis for gene discovery.






Similar content being viewed by others
References
Ahfock D, Wood I, Stephen S, Cavanagh CR, Huang BE (2014) Characterizing uncertainty in high-density maps from multiparental populations. Genetics 198:117–128
Araus JL, Cairns JE (2014) Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci 19:52–61
Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA et al (2011) Genetic analysis of complex traits in the emerging collaborative cross. Genome Res 21:1213–1222
Bailey DW (1971) Recombinant-inbred strains: an aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11:325–327
Bandillo N, Raghavan C, Muyco PA, Sevilla MAL, Lobina IT et al (2013) Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6:11
Bardol N, Ventelon M, Mangin B, Jasson S, Loywick V et al (2013) Combined linkage and linkage disequilibrium QTL mapping in multiple families of maize (Zea mays L.) line crosses highlights complementarities between models based on parental haplotype and single locus polymorphism. Theor Appl Genet 126:2717–2736
Bink MCAM, Boer MP, Ter Braak CJF, Jansen J, Voorrips RE et al (2008) Bayesian analysis of complex traits in pedigreed plant populations. Euphytica 161:85–96
Blakeslee AF, Belling J, Farnham ME, Bergner AD (1922) A haploid mutant in the Jimson weed, “Datura Stramonium”. Science 55:646–647
Bottomly D, Ferris MT, Aicher LD, Rosenzweig E, Whitmore A et al (2012) Expression quantitative trait loci for extreme host response to influenza A in pre-Collaborative Cross mice. G3 2:213–221
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brim CA (1966) A modified pedigree method of selection in soybeans. Crop Sci 6:220
Broman K (2005) The genomes of recombinant inbred lines. Genetics 169:1133–1146
Broman KW (2012) Genotype probabilities at intermediate generations in the construction of recombinant inbred lines. Genetics 190:403–412
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Buet C, Dubreuil P, Tixier M-H, Durantin K, Praud S et al (2013) The molecular characterization of a MAGIC population reveals high potential for gene discovery. MaizeGDB proceedings
Butler D (2009) asreml: asreml() fits the linear mixed model. R package version 3.0. http://www.vsni.co.uk
Cavanagh C, Morell M, Mackay I, Powell W (2008) From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Op Plant Biol 11:215–221
Cavanagh C, Chao S, Wang S, Huang BE, Stephen S et al (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci 110:8057–8062
Collaborative Cross Consortium (2012) The genome architecture of the collaborative cross mouse genetic reference population. Genetics 190:389–401
Complex Trait Consortium (2004) The collaborative cross, a community resource for the genetic analysis of complex traits. Nat Genet 36:1133–1137
Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF (2013) Genetic incompatibilities are widespread within species. Nature 504:135–137
Cullis BR, Smith AB, Coombes NE (2006) On the design of early generation variety trials with correlated data. J Agric Biol Environ Stat 11:381–393
Darvasi A, Soller M (1995) Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141:1199–1207
Das S, Zijdenbos AP, Harlap J, Vins D, Evans AC (2011) LORIS: a web-based data management system for multi-center studies. Front Neuroinform 5:37
Demarest K, Koyner J, McCaughran J Jr, Cipp L, Hitzemann R (2001) Further characterization and high- resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav Genet 31:79–91
Durrant C, Mott R (2010) Bayesian quantitative trait locus mapping using inferred haplotypes. Genetics 184:839–852
Durrant C, Swertz MA, Alberts R, Arends D, Moller S et al (2012) Bioinformatics tools and database resources for systems genetics analysis in mice—a short review and an evaluation of future needs. Brief Bioinform 13:135–142
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K et al (2011) A robust, simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS One 6(5):e19379
Esch E, Szymaniak JM, Yates H, Pawlowski WP, Buckler ES (2007) Using crossover breakpoints in recombinant inbred lines to identify quantitative trait loci controlling the global recombination frequency. Genetics 177:1851–1858
Forster BP, Bors-Heberle E, Kasha KJ, Touraev A (2007) The resurgence of haploids in higher plants. Trends Plant Sci 12:368–375
Furbank RT, Tester M (2011) Phenomics—technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16:635–644
Gan X, Stegle O, Behr J, Steffen JG, Drewe P et al (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477:419–423
Gaur PM, Jukanti AK, Varshney RK (2012) Impact of genomic technologies on chickpea breeding strategies. Agronomy 2:199–221
Giraud H, Lehermeier C, Bauer E, Falque M, Segura V et al (2014) Linkage disequilibrium with linkage analysis of multiline crosses reveals different multiallelic QTL for hybrid performance in the Flint and Dent heterotic groups for maize. Genetics 198:1717–1734
Goulden CH (1939) Problems in plant selection. In: Proceedings of the Seventh International Genetics Congress. Cambridge University Press, pp 132–133
Green PJ (1995) Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82:711–732
Gyenesei A, Moody J, Semple CAM, Haley CS, Wei W-H (2012) High throughput analysis of epistasis in genome-wide association studies with BiForce. Bioinformatics 28:1957–1964
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324
Harushima Y, Yano M, Shomura A, Sato M, Shimano T et al (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148:479–494
Hemani G, Theocharidis A, Wei W, Haley C (2011) EpiGPU: exhaustive pairwise epistasis scans parallelized on consumer level graphics cards. Bioinformatics 27:1462–1465
Hickey JM, Gorjanc G, Hearne S, Huang BE (2014) AlphaMPSim: flexible simulation of multi-parent crosses. Bioinformatics 30:2686–2688
Howe D, Costanzo M, Fey P, Gojobori T, Hannick L et al (2008) Big data: the future of biocuration. Nature 455:47–50
Huang BE, George AW (2011) R/mpMap: a computational platform for the genetic analysis of multi-parent recombinant inbred lines. Bioinformatics 27:727–729
Huang X, Feng Q, Qian Q, Zhao Q, Wang L et al (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Huang X, Paulo M-J, Boer M, Effgen S, Keizer P et al (2011) Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. PNAS 108(11):4488–4493
Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ et al (2012) A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J 10:826–839
Huang BE, Clifford D, Cavanagh C (2013) Selecting subsets of genotyped experimental populations for phenotyping to maximize genetic diversity. Theor Appl Genet 126:379–388
Huang BE, Raghavan C, Mauleon R, Broman KW, Leung H (2014) Imputation of low-coverage genotyping-by-sequencing in multi-parental crosses. Genetics 197:401–404
Jansen RC (1994) Controlling the type I and type II errors in mapping quantitative trait loci. Genetics 138:871–881
Kao CH, Zeng ZB, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216
Kass RE (1993) Bayes factors in practice. Statistician 42:551–560
Kass RE, Raftery AE (1995) Bayes factors. JASA 90:773–795
King EG, Macdonald SJ, Long AD (2012a) Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics 191:935–949
King EG, Merkes CM, McNeil CL, Hoofer SR, Sen S et al (2012b) Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res 22:1558–1566
King EG, Sanderson BJ, McNeil CL, Long AD, Macdonald SJ (2014) Genetic dissection of the Drosophila melanogaster female head transcriptome reveals widespread allelic heterogeneity. PLoS Genet 10(5):e1004322
Klasen JR, Piepho H-P, Stich B (2012) QTL detection power of multi-parental RIL populations in Arabidopsis thaliana. Heredity 108:626–632
Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM et al (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5(7):e1000551
Lai K, Lorenc MT, Edwards D (2012) Genomic databases for crop improvement. Agronomy 2:62–73
Leroux D, Rahmani A, Jasson S, Ventelon M, Louis F et al (2014) Clusthaplo: a plug-in for MCQTL to enhance QTL detection using ancestral alleles in multi-cross design. Theor Appl Genet 127:921–933
Mace ES, Hunt CH, Jordan DR (2013) Supermodels: sorghum and maize provide mutual insight into the genetics of flowering time. Theor Appl Genet 126:1377–1395
Mackay IJ, Bansept-Basler P, Barber T, Bentley AR, Cockram J et al (2014) An eight-parent Multiparent Advanced Generation Inter-Cross population for winter-sown wheat: creation, properties and validation. G3 4:1603–1610
Malosetti M, van Eeuwijk DA, Boer MP, Casas AM, Elia M et al (2011) Gene and QTL detection in a three-way barley cross under selection by a mixed model with kinship information using SNPs. Theor Appl Genet 122:1605–1616
Malosetti M, Ribaut J-M, van Eeuwijk FA (2013) The statistical analysis of multi-environment data: modelling genotype-by-environment interaction and its genetic basis. Front Physiol 4:44
Maluszynski M, Kasha KJ, Szareiko I (2003) Published doubled haploid protocols in plant species. In: Doubled haploid production in crop plants, a manual. Kluwer Academic Publishers, Dordecht, pp 309–335
McClearn GE, Wilson JR, Meredith W (1970) The use of isogenic and heterogenic mouse stocks in behavioral research. In: Lindzey G, Thiessen D (eds) Contributions to behavior-genetic analysis: the mouse as a prototype. Appleton Century Crofts, New York, pp 3–22
McMullen MD, Kresovich S, Villeda HS, Bradbury PJ, Li H et al (2009) Genetic properties of the maize nested association mapping population. Science 325:737–740
Meuwissen TH, Goddard ME (2001) Prediction of identity by descent probabilities from marker-haplotypes. Genet Sel Evol 33:605
Mohring J, Piepho H-P (2009) Comparison of weighting in two-stage analyses of series of experiments. Crop Sci 39:1977–1988
Montes JM, Melchinger AE, Reif JC (2007) Novel throughput phenotyping platforms in plant genetic studies. Trends Plant Sci 12:433–436
Mott R, Talbot CJ, Turri MG, Collins AC, Flint J (2000) A new method for fine-mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA 97:12649–12654
Pascual L, Desplat N, Huang BE, Desgroux A, Bruguier L et al (2015) Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol J (in press)
Pea G, Dell’Acqua M, Hlaing ALL, Pe ME (2013) From mice to maize: a multiparental population for fine mapping in Zea mays. MAGIC Populations Workshop. http://openwetware.org/images/e/e6/MatteoDellAcqua_MaizePoster.pdf
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ram R, Mehta M, Balmer L, Gatti DM, Morahan G (2014) Rapid identification of major effect genes using the Collaborative Cross. Genetics 198:75–86
Rebetzke GJ, Verbyla AP, Verbyla KL, Morell MK, Cavanagh CR (2014) Use of a large multiparent wheat mapping population in genomic dissection of coleoptile and seedling growth. Plant Biotechnol J 12:219–230
Sannemann W, Huang BE, Mathew B, Léon J (2015) Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol Breeding 35:86
Schmitt CP, Burchinal M (2011) Data management practices for collaborative research. Front Psychiatry 2:47
Schnaithmann F, Kopahnke D, Pillen K (2014) A first step toward the development of a barley NAM population and its utilization to detect QTLs conferring leaf rust seedling resistance. Theor Appl Genet 127:1513–1525
Scutari M, Howell P, Balding DJ, Mackay IJ (2014) Multiple quantitative trait analysis using Bayesian networks. Genetics 198:129–137
Smith AB, Lim P, Cullis BR (2006) The design and analysis of multi-phase plant breeding experiments. J Agric Sci Camb 144:393–409
Smith AB, Thompson R, Butler DC, Cullis BR (2011) The design and analysis of variety trials using mixtures of composite and individual plot samples. J Royal Stat Soc C 60:437–455
Smith AB, Butler DG, Cavanagh CR, Cullis BR (2015) Multi-phase variety trials using both composite and individual replicate samples: a model-based design approach. J Agric Sci Camb (in press)
Stich B (2009) Comparison of mating designs for establishing Nested Association Mapping populations in maize and Arabidopsis thaliana. Genetics 183:1525–1534
Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R et al (2012) High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190:437–447
Thépot S, Restoux G, Goldringer I, Hospital F, Gouache D, Mackay I, Enjalbert J (2015) Efficiently tracking selection in a multiparental population: the case of earliness in wheat. Genetics 199:609–623
Valdar W, Flint J, Mott R (2006) Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172:1783–1797
Valdar W, Holmes CC, Mott R, Flint J (2009) Mapping in structured populations by resample model averaging. Genetics 182:1263–1277
van Eeuwijk FA, Bink MC, Chenu K, Chapman SC (2010) Detection and use of QTL for complex traits in multiple environments. Curr Opin Plant Biol 13:193–205
Verbyla AP, Cullis BR (2012) Multivariate whole genome average interval mapping: QTL analysis for multiple traits and/or environments. Theor Appl Genet 125:933–953
Verbyla AP, Cullis BR, Thompson R (2007) The analysis of QTL by simultaneous use of the full linkage map. Theor Appl Genet 116:95–111
Verbyla AP, George AW, Cavanagh CR, Verbyla KL (2014a) Whole genome QTL analysis for MAGIC. Theor Appl Genet 127:1753–1770
Verbyla AP, Cavanagh CR, Verbyla KL (2014b) Whole genome analysis of multi-environment or multi-trait QTL in MAGIC G3(4):1569–1584
Wang J, de Villena FP, Lawson HA, Cheverud JM, Churchill GA et al (2012) Imputation of single-nucleotide polymorphisms in inbred mice using local phylogeny. Genetics 190:449–458
Wang S, Wong D, Forrest K, Allen A, Chao S et al (2014) Characterization of polyploidy wheat genomic diversity using the high-density 90,000 SNP array. Plant Biotechnol J 12:787–796
Xu S (1996) Mapping quantitative trait loci using four-way crosses. Genet Res 68:175–181
Yalcin B, Flint J, Mott R (2005) Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics 171:673–681
Yamamoto E, Iwata H, Tanabata T, Mizobuchi R, Yonemaru J et al (2014) Effect of advanced intercrossing on genome structure and on the power to detect linked quantitative trait loci in a multi-parent population: a simulation study in rice. BMC Genet 15:50
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Acknowledgments
Many thanks to three anonymous reviewers for their helpful suggestions. Dr. Huang is the recipient of an Australian Research Council Discovery Early Career Researcher Award (Project Number DE120101127).
Conflict of interest
No authors have any conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. H. Geiger.
Rights and permissions
About this article
Cite this article
Huang, B.E., Verbyla, K.L., Verbyla, A.P. et al. MAGIC populations in crops: current status and future prospects. Theor Appl Genet 128, 999–1017 (2015). https://doi.org/10.1007/s00122-015-2506-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2506-0