Abstract
In many cases, an unknown to an investigator is actually known in the chemical literature, a reference database, or an internet resource. We refer to these types of compounds as “known unknowns.” ChemSpider is a very valuable internet database of known compounds useful in the identification of these types of compounds in commercial, environmental, forensic, and natural product samples. The database contains over 26 million entries from hundreds of data sources and is provided as a free resource to the community. Accurate mass mass spectrometry data is used to query the database by either elemental composition or a monoisotopic mass. Searching by elemental composition is the preferred approach. However, it is often difficult to determine a unique elemental composition for compounds with molecular weights greater than 600 Da. In these cases, searching by the monoisotopic mass is advantageous. In either case, the search results are refined by sorting the number of references associated with each compound in descending order. This raises the most useful candidates to the top of the list for further evaluation. These approaches were shown to be successful in identifying “known unknowns” noted in our laboratory and for compounds of interest to others.
Similar content being viewed by others
1 Introduction
We have previously demonstrated [1] that searching the Chemical Abstracts Service (CAS) Registry employing accurate mass mass spectrometry data is a very useful approach for the identification of “known unknowns.” We define a “known unknown” as a compound which is unknown to the investigator but is known in the chemical literature, a reference database, or an internet resource.
There is a particular need for additional approaches for the identification of “known unknowns” found in liquid chromatography/mass spectrometry (LC/MS) analyses because the availability of computer searchable collision-induced dissociation (CID) mass spectral databases is limited for LC/MS [1] compared with that of electron ionization (EI) mass spectral databases. We have found that searching “spectraless” databases such as the CAS Registry [1] by elemental compositions or molecular weights, then sorting the hit list in descending order by the number of associated references to be very effective in the identification of “known unknowns.” The most likely candidates are normally brought to the top of the list where they can be further scrutinized by employing additional data.
ChemSpider is another very large “spectraless” database that can be searched by elemental composition, molecular weight, or monoisotopic mass, and it is provided as a free resource to the community. ChemSpider contains more than 26 million entries compared with the CAS Registry that contains more than 62 million substances. In our current work, the ChemSpider interface was modified [2] such that the initial search results can be sorted by the number of references associated with an entry. The approach was then evaluated with a wide variety of compounds and compared with previous results obtained with the CAS Registry [1].
2 Experimental
2.1 ChemSpider Software Modifications
Several changes were made in the ChemSpider software interface to facilitate our studies [2]. The most important change was the ability to sort the initial search results in descending order by the number of data sources or associated references. The references in ChemSpider originate from the SureChem patent database (>20 million), PubMed (>20 million articles), and the content of the Royal Society of Chemistry publishing database. In our work, sorting the “# of references” column was found to be the most useful. Many screen displays of the software annotated with examples from our laboratory are shown in the Electronic Supplementary Material.
Two other changes allowed more convenient data entry for searching by the user. The first change was the ability for the user to enter the m/z value directly for a charged species and then select the type of ionic species from a pull-down (drop-down) menu. Examples of typical types of ionic species in the menu include [M + Na]+, [M + NH4]+, [M – H]–, etc. The m/z value entered is then automatically adjusted by the program before searching the monoisotopic mass of the neutral species as it would appear in the ChemSpider database.
The second change was the ability for the entered m/z value of the charged species to be corrected for the mass of an electron. Some manufacturers’ data systems do not properly calculate the m/z values of ions for the mass of an electron [3, 4]. The errors normally cancel within the manufacturers’ elemental composition programs because the reference calibration tables are also not corrected. However, all data exported into other applications for further data processing should be corrected.
2.2 Acquisition of Accurate Mass Data
The experimental detail, including calibrations, typical chromatographic separations, and sample preparations were previously described in detail [1]. Briefly, the accurate mass electrospray LC/MS/UV-Vis (ultraviolet-visible) data were obtained on a LCT time-of-flight mass spectrometer (Waters Corporation, Milford, MA, USA) equipped with a LockSpray secondary ESI probe. An 1100 Series liquid chromatograph with autosampler, degasser, and UV-Vis diode array spectrophotometer (Agilent Technologies, Santa Clara, CA, USA) was employed for the separations in the reversed phase mode. The UV-Vis spectral data is very useful in locating particular classes of compounds (UV absorbers, dyes, etc.) and in confirming the identity of compounds by comparison to reference spectra from standards, literature references, or even internet sources.
Elemental compositions were generated from monoisotopic masses utilizing the Waters Elemental Composition Program within MassLynx ver. 4.1 software, which included i-FIT software for numerically ranking the observed isotopic pattern to the theoretical ones. Our LCT accurate mass measurements typically yielded a standard deviation of 5 ppm. Thus, windows of ±18 ppm (slightly greater than three standard deviations) were employed in examples from our laboratory to insure inclusions of all reasonable candidates for determining elemental compositions or for searching ChemSpider by monoisotopic masses. In literature studies, windows of approximately ±5 ppm were employed because many currently available accurate mass time-of-flight instruments yield standard deviations approaching 1 ppm for mass measurements.
3 Results and Discussion
3.1 Descriptions of Two Approaches
The ability to search by either elemental composition or monoisotopic mass is very valuable for the identification of “known unknowns” using accurate mass mass spectrometry data. In the work described within this article, the ChemSpider user interface was modified to permit the sorting of either the elemental composition or monoisotopic mass search results by the number of associated references. The results, sorted in descending order, normally bring the most useful entries to the top of the list. These candidate structures are then scrutinized [1] by other available data such as in-source CID spectra, UV-Vis spectra, GC/MS data, number of exchangeable protons, NMR data, etc. to ultimately obtain the identification of the compound of interest. For critical identifications, a standard of the material is normally obtained and its LC retention time, UV-Vis spectrum, and in-source CID spectra are compared with those of the unknown.
A very similar approach [1] was demonstrated to be very useful utilizing the CAS Registry of more than 62 million substances, a fee-based service, which is searched by either STN Express or the web-based version of SciFinder. The CAS Registry can only be searched by elemental composition, molecular weight, or nominal molecular weight, but not monoisotopic mass. The ability to search by the monoisotopic mass is much more useful because the standard deviation for its determination is much lower than that for the molecular weight. In addition, the monoisotopic mass is calculated by all manufacturers’ data systems, whereas the molecular weight must be manually calculated by the user.
One significant advantage of searching the CAS Registry versus the ChemSpider database is the ability of the former to search its more than 34 million document records associated with an elemental composition by key words [1]. Only very minimal sample history is required to quickly obtain useful candidates for tentative identifications by this approach. This capability can be especially useful in identifying more obscure “known unknowns” with fewer associated references. This capability is not currently available with ChemSpider.
3.2 Evaluation of the Two Approaches with Literature Examples
A group of 90 compounds was assembled from literature sources [5–8], internet sites, and American Society of Mass Spectrometry Conference presentations to evaluate the two approaches in ChemSpider. The results are summarized in Tables 1 and 2. Searching the ChemSpider database by elemental compositions, then sorting in descending order by the number of overall references yielded more target compounds highly ranked compared with the same approach using monoisotopic masses. This is to be expected because the number of overall candidates was less when searching by elemental composition (mean = 513, median = 310) compared with monoisotopic mass (mean = 800, median = 740). However, the overall number with rankings less than or equal to 5 was acceptable in both cases. Thus, searching by elemental composition is the preferred approach, but searching by a monoisotopic mass is also a reasonable approach when a unique elemental composition cannot be readily determined.
The same 90 compounds were previously evaluated with the CAS Registry searching by elemental compositions [1] using the web-based version of SciFinder. Similar results (see bottom of Table 1) were obtained using either ChemSpider or the CAS registry as databases for these limited number of test compounds. The CAS Registry currently cannot be searched by monoisotopic mass [1]; therefore, no direct comparison can be made in Table 2 for the results obtained with ChemSpider.
3.3 Example from Our Laboratory for UV Stabilizer Identification in a Commercial Polymer
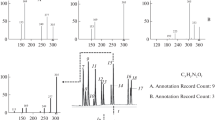
An unknown additive, noted in a commercial polymer sample, was characterized by accurate mass LC/MS. The accurate mass data was used in conjunction with isotopic abundance information to obtain an elemental composition of C22H29N3O. ChemSpider was searched by the elemental composition and 1135 hits were found, which were sorted in descending order by the number of overall associated references. The top candidate was Tinuvin 328. The proposed structure was consistent with the accurate mass in-source CID spectrum (Scheme 1), which showed two very significant losses of C5H10 from the protonated molecule, [M + H]+, and the presence of a C5H +11 ion at a m/z value of 71.085.
The confidence afforded by the initial data allowed the identity to be reported to the customer. At a later date, the identification of the additive was confirmed by comparison of its LC retention time, UV-Vis spectrum, and in-source CID mass spectra to those of a purchased reference sample.
ChemSpider was also searched for protonated molecules with m/z 352.239 using a m/z window of ±18 ppm. There were 1459 hits and Tinuvin 328 was still the top candidate. The mass precision of our older instrumentation is relatively large, but newer time-of-flight instrumentation afford much better mass precision, which would return a smaller number of candidates from the search. Screen displays from the ChemSpider interface for the identification of Tinuvin 328 by both approaches are shown in the Electronic Supplementary Material.
3.4 Advantage of Searching Higher MW Compounds by Monoisotopic Mass Data
It is often difficult to determine a unique elemental composition for “known unknowns” with molecular weights greater than 600 Da [9, 10]. Either there are two or more possible elemental compositions for the unknown or one is inadvertently excluded if the somewhat subjective user settings are set too narrow for elements present, range of elements, double bond equivalents, etc. In theory, the number of elemental compositions increases dramatically as the molecular weight increases. However, in practice, the number of elemental compositions at molecular weights greater than 600 Da decreases dramatically in both ChemSpider (see Figure 1) and CAS Registry databases [1].
Therefore, it is much more reasonable to search for “known unknowns” in these databases using monoisotopic mass instead of struggling to determine a unique elemental composition for searching. The rankings noted by number of references for several higher molecular weight compounds noted in the literature [9, 10] are compared with elemental composition searches versus monoisotopic mass searches in Table 3. There is essentially no significant penalty noted for searching by monoisotopic mass instead of elemental composition for this limited number of examples. Of course, ex-post facto, it is still extremely important to compare the isotopic abundances of the candidate structures from ChemSpider to those of the unknown. The ability to calculate, rank, and compare theoretical isotopic abundances for elemental compositions to those of observed ones is normally a standard option in most mass spectrometry manufacturers’ software. The CAS and ChemSpider identification numbers for compounds in Table 3 are included in the Electronic Supplementary Material.
3.5 Example from Our Laboratory for the Identification of a Higher Molecular Weight Antioxidant in a Commercial Polymer
An unknown additive, noted in a commercial polymer sample, was characterized by accurate mass LC/MS. The ammonium adduct, [M + NH4]+, of the component was observed at a m/z value of 801.558. The observed ion was confirmed to be an ammonium adduct because at higher in-source CID energies the ammonium adduct intensity was reduced and the intensity of the sodium adduct was noted to increase. This increase in absolute intensity of the sodium adduct and corresponding decrease in that of the ammonium adduct is routinely noted in our laboratory for in-source CID [1] and a typical example is shown in the Electronic Supplementary Material.
ChemSpider was searched by the monoisotopic mass for the ammonium adduct with a mass window of ±18 ppm (see Electronic Supplementary Material) and 23 candidates were obtained. The top candidate was Goodrite 3114. Only two of the 23 candidates had isotopic abundances consistent with that of the unknown. Of these two, only the in-source CID mass spectrum (Scheme 2) of Goodrite 3114 was consistent with that for the unknown.
At a later date, the LC retention time, UV-Vis spectrum, and in-source CID mass spectra of a commercial sample were shown to be identical to those of the unknown.
3.6 Future Enhancements to Improve Productivity
ChemSpider currently supports a web application program interface (API) via web services (http://www.chemspider.com/MassSpecAPI.asmx) for batch queries by external vendors’ programs. Several companies, including Bruker Daltonics, Thermo Scientific, Waters, and Agilent Technologies, integrate their data processing programs with ChemSpider. Users who integrate using the web services, including these vendors, use a token supplied by ChemSpider to authenticate their identity to the web service. Currently there are no similar capabilities for such queries of the CAS Registry using either SciFinder or STN Express.
Some changes in both the current API and the vendors’ programs would be required to utilize associated references and comparison of isotopic abundances to facilitate increases in data processing speeds. When searching by monoisotopic mass, the isotopic abundances of the candidates should then be calculated in the vendor’s data system and compared with the observed isotopic abundance of the unknown and sorted in descending order by either the number of references or the fit of the isotopic abundances to those of the unknown. This would be particularly useful for “known unknowns” with molecular weights greater than 600 Da, but would be also beneficial for lower molecular weight compounds. Alternatively, the ChemSpider software could be enhanced to perform the calculations of the isotopic abundances of the candidates for comparison to the unknowns.
Even with the above changes, excessive time would still be needed to manually compare the observed CID spectrum of an unknown with fragments expected by the user from the candidates’ structures. If in silico CID spectra, i.e., theoretical fragmentation with predicted abundances, could be calculated [11, 12] and ranked for the candidate structures from either ChemSpider or SciFinder SDF (Structure Data Format) files [13], further data processing speeds would be realized. The results of the numeric structure ranking, isotopic abundance, and number of references could then be more quickly examined by the user to yield tentative identifications.
Both ChemSpider and the web-based SciFinder are able to export structures for the candidate compounds in SDF. The web-based version SciFinder permits the export of up to 500 structures in an SDF file. There is no limit to the number of structures that can be exported in the ChemSpider API and a limit of 10,000 in the web interface.
3.7 Charged Species in ChemSpider
More development work needs to be performed to facilitate the searching of charged species by either elemental composition or monoisotopic mass. Sodium benzoate illustrates the current state of affairs for organic anions. Its elemental composition and monoisotopic mass are listed in ChemSpider as C7H5NaO2 and 144.018724, respectively. It would be more useful to parse the elemental composition as C7H6O2.Na (neutral benzoic acid to left of period, associated cation to the right) then list the elemental composition and monoisotopic mass for searching as C7H6O2 and 122.037, respectively. This is the format employed in the CAS Registry [1]. In reverse phase chromatography electrospray mass spectrometry, the organic anions elute as their free acid form and are normally detected as [M – H]– ions in the negative ion mode. Thus, their identity is independent of their associated cation.
A simple example of an organic cation is N,N,N-trimethyl-N-benzylammonium acetate, whose elemental composition and monoisotopic mass are listed as C12H19NO2 and 209.1216, respectively, in ChemSpider. It would be more beneficial to parse this type of organic cation as C10H16N.C2H3O2, then list the elemental composition and monoisotopic mass for searching, respectively, as C10H16N and 150.1277.
Amphoteric (inner salt, zwitterionic) species are listed in the database with no modifications. For example, (CH3)3 N+CH2CO –2 , trimethylglycine, is listed as C5H11NO2 with monoisotopic mass of 117.079. This type of compound would yield [M + H]+ and [M + acetate]– ions, respectively, in positive and negative ion electrospray analyses using acetate in the LC eluent. Thus the elemental composition for the species would need to be corrected before searching.
4 Conclusions
Modifications in the ChemSpider interface to sort elemental composition and monoisotopic mass search results by the number of associated references in descending order offered significantly improved capabilities for the identification of “known unknowns” using accurate mass mass spectrometry data. Other changes were made to allow easier input of data by users to specify the type of ion adduct and to correct the monoisotopic mass for the mass of an electron. Further enhancements are still needed to improve the overall productivity of the process and to enable the searching of charged species.
The elemental composition search is the preferred one in the lower molecular weight range (200–600 Da), but even the monoisotopic mass search with reasonable error windows is viable in this range. Monoisotopic mass searching for compounds with a molecular weight >600 Da is preferred when it is difficult to determine a unique elemental composition. The resulting candidates from the monoisotopic mass search are then ranked by comparing their calculated isotopic abundances by the manufacturers’ isotope abundance programs to that of the unknown.
The ability to search monoisotopic mass in ChemSpider is an important function, which is absent in search engines for the CAS Registry. However, CAS Registry searches can be further refined by key words, which can be particularly useful for more obscure “known unknowns” with few associated references. This option is not currently available in ChemSpider. The ability to search by both databases is very desirable depending on the problem at hand when costs are not a limitation. ChemSpider is provided at no cost to the community, but there is a fee associated with the utilization of the CAS Registry.
In all cases, other data such as sample history, UV-Vis data, types of ions observed, exchangeable protons, CID spectra, etc. are needed to scrutinize the candidate structures from the elemental composition and monoisotopic mass search results. For ultimate confirmation of structure, analysis of a standard material under identical conditions is always desirable.
References
Little, J.L., Cleven, C.D., Brown, S.D.: Identification of “Known Unknowns” utilizing accurate mass data and chemical abstracts service databases. J. Am. Soc. Mass Spectrom. 22, 348–359 (2011)
Little, J.L., Williams, A.J., Tkachenko, V.: Pshenichnov, A: Accurate Mass Measurements: Identifying “Known Unknowns” Using ChemSpider. Proceedings of the ASMS, Denver, CO (2010)
Mamer, O.A., Lesimple, A.: Common shortcomings in computer programs calculating molecular weights. J. Am. Soc. Mass Spectrom. 14, 626 (2004)
Ferrer, I., Thurman, E.M.: Importance of the electron mass in the calculations of exact mass by time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 21, 2538–2539 (2007)
Hogenboom, A.C., van Leerdam, J.A., de Voogt, P.: Accurate mass screening and identification of emerging contaminants in environmental samples by liquid chromatography-hybrid linear ion trap orbitrap mass spectrometry. J. Chromatogr. A 1216, 510–519 (2009)
Liao, W., Draper, W.M., Perera, S.K.: Identification of unknowns in atmospheric pressure ionization mass spectrometry using a mass to structure search engine. Anal. Chem. 80, 7765–7777 (2008)
Garcia-Reyes, J.F., Ferrer, I., Thurman, E.M., Molina-Diaz, A., Fernandez-Alba, A.R.: Searching for non-target chlorinated pesticides in food by liquid chromatography/time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 19, 2780–2788 (2005)
Ojanperä, S., Pelander, A., Peizing, M., Krebs, I., Vuori, E., Ojanperä, I.: Isotopic pattern and accurate mass determination in urine drug screening by liquid chromatography/time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 20, 1161–1167 (2006)
Erve, J.C., Gu, M., Wang, Y., DeMaio, W., Talaat, R.E.: Spectral accuracy of molecular ions in an LTQ/Orbitrap mass spectrometer and implications for elemental composition determination. J. Am. Soc. Mass Spectrom. 20, 2058–2069 (2009)
Lohmann, W.; Decker, P; Barsch, A: Accurate Elemental Composition Determination and Identification of Molecules with >1100 m/z with UHR-TOF. Bruker Application Note # ET-23 (2011).
Sweeney, D.L.: Small molecules as mathematical partitions. Anal. Chem. 75, 5362–5373 (2003)
Hill, A.W., Mortishire-Smith, J.: Automated assignment of high-resolution collisionally activated dissociation mass spectra using a systematic bond disconnection approach. Rapid Comm. Mass Spectrom. 19, 3111–3118 (2005)
Dalby, A., Nourse, J.G., Hounshel, W.D., Gushurst, A.K.I., Grier, D.L., Leland, B.A., Laufer, J.: Description of several chemical structure file formats used by computer programs developed at molecular design Ltd. J. Chem. Inf. Comput. Sci. 32, 244–255 (1992)
Acknowledgments
The authors gratefully recognize Curt Cleven from Eastman Chemical Company, Mike Scott from Agilent Technologies, Inc., and Jim Lekander from Waters Corporation for their help and advice. They also thank Bill Tindall and Kent Morrill (retirees from Eastman Chemical Company) for their initial work on “spectraless” databases created from the Toxic Substances Control Act (TSCA) and the Eastman Corporate Plant Material databases.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 315 kb)
Rights and permissions
About this article
Cite this article
Little, J.L., Williams, A.J., Pshenichnov, A. et al. Identification of “Known Unknowns” Utilizing Accurate Mass Data and ChemSpider. J. Am. Soc. Mass Spectrom. 23, 179–185 (2012). https://doi.org/10.1007/s13361-011-0265-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-011-0265-y
Key words
- Accurate mass mass spectrometry
- Unknown identification
- Known unknowns
- Electrospray
- Liquid chromatography/mass spectrometry
- LC/MS
- ChemSpider
- CAS registry
- SciFinder
- STN express
- Monoisotopic mass
- Molecular formulae
- Time-of-flight mass spectrometry
- TOF MS
- Collision-induced dissociation
- CID
- MS/MS
- Elemental composition
- Molecular formula
- Chemical Abstracts Service
- Isotopic abundances
- Tandem mass spectrometry
- molecular weight
- In-source CID