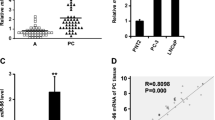
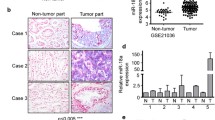
Abstract
In developed countries, prostate cancer (PC) is the neoplasia more frequently diagnosed in men. The signaling pathway induced by the transforming growth factor β1 (TGFβ1) has an important role in cell growth, differentiation, and development, the downregulation of this pathway being associated with cancer development. In PC, the activation of this signaling pathway is lost, resulting in favoring of tumor growth, proliferation, and evasion of apoptosis. Several studies have shown that microRNAs (miRNAs), small non-coding RNA, are closely associated with the development, invasion, and metastasis, suggesting that they have a critical role in cancer development. Recently, Smad proteins, the signal transducers of the TGFβ1 signaling pathway, were found to regulate miRNA expression, through both transcriptional and posttranscriptional mechanisms. In this review, we summarize the mechanisms underlying Smad-mediated regulation of miRNA biogenesis and the effects on cancer development, particularly in PC. We identify that TGFβ1-related miR-143, miR-145, miR-146a, and miR-199a may have a key role in the development of prostate cancer metastasis and the restoration of their expression may be a promising therapeutic strategy for PC treatment.




Similar content being viewed by others
References
Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–81.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: Globocan 2008. Int J Cancer. 2010;127:2893–917.
Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, Brawley O, et al. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61:1079–92.
Semenas J, Allegrucci C, Boorjian SA, Mongan NP, Persson JL. Overcoming drug resistance and treating advanced prostate cancer. Curr Drug Targets. 2012;13:1308–23.
Kumar V, Cotran R, Collins T. Robbins and Cotran pathologic basis of disease, ed 7. 2005.
Chen YC, Page JH, Chen R, Giovannucci E. Family history of prostate cancer and breast cancer and the risk of prostate cancer in the PSA era. Prostate. 2008;68:1582–91.
Heidenreich A, Bellmunt J, Bolla M, Joniau S, Mason M, Matveev V, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol. 2011;59:61–71.
Houlgatte A, Vincendeau S, Desfemmes F, Ramirez J, Benoist N, Bensalah K, et al. Use of [-2] pro PSA and phi index for early detection of prostate cancer: A prospective of 452 patients. Prog Urol J Assoc Fr Urol Soc Fr Urol. 2012;22:279–83.
Stephan C, Vincendeau S, Houlgatte A, Cammann H, Jung K, Semjonow A. Multicenter evaluation of [-2]proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clin Chem. 2013;59:306–14.
Zhang J, Zhao H, Gao Y, Zhang W. Secretory miRNAs as novel cancer biomarkers. Biochim Biophys Acta. 1826;2012:32–43.
Hao Y, Zhao Y, Zhao X, He C, Pang X, Wu TC, et al. Improvement of prostate cancer detection by integrating the PSA test with miRNA expression profiling. Cancer Investig. 2011;29:318–24.
Fontenete S, Silva J, Teixeira AL, Ribeiro R, Bastos E, Pina F, et al. Controversies in using urine samples for prostate cancer detection: PSA and PCA3 expression analysis. Int Braz J Urol Off J Braz Soc Urol. 2011;37:719–26.
Nam RK, Zhang W, Siminovitch K, Shlien A, Kattan MW, Klotz LH, et al. New variants at 10q26 and 15q21 are associated with aggressive prostate cancer in a genome-wide association study from a prostate biopsy screening cohort. Cancer Biol Ther. 2011;12:997–1004.
Hirata H, Hinoda Y, Kikuno N, Kawamoto K, Dahiya AV, Suehiro Y, et al. Cxcl12 g801a polymorphism is a risk factor for sporadic prostate cancer susceptibility. Clin Cancer Res. 2007;13:5056–62.
Wang SK, Wang ZZ, Huang YF. [Advances in researches on the relationship between single nucleotide polymorphism and prostate cancer]. Zhonghua nan ke xue =. Natl J Androl. 2005;11:605–10.
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326–31.
Hou X, Flaig TW. Redefining hormone sensitive disease in advanced prostate cancer. Adv Urol. 2012;2012:978531.
Nicolas Mottet JB. Michel Bolla, Steven Joniau, Malcolm Masone, et al. EAU guidelines on prostate cancer. Part II: treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol. 2011;59:572–83.
Silva FCd (ed) Recomendações clínicas no tratamento do carcinoma da próstata, Lisboa, 2013, pp 226.
Saraon P, Jarvi K, Diamandis EP. Molecular alterations during progression of prostate cancer to androgen independence. Clin Chem. 2011;57:1366–75.
Attar RM, Takimoto CH, Gottardis MM. Castration-resistant prostate cancer: locking up the molecular escape routes. Clin Cancer Res. 2009;15:3251–5.
Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45.
Kavleen Sikand SB, Girish C. Shukla. MicroRNAs and androgen receptor 3′ untranslated region: a missing link in castration-resistant prostate cancer? Mol Cell Pharmacol. 2011;3:107–13.
Keith F. Decker DZ, Yuhong He, Tamara Bowman, John R. Edwards and Li Jia. Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Res. 2012:1–15.
Lamont KR, Tindall DJ. Minireview: alternative activation pathways for the androgen receptor in prostate cancer. Mol Endocrinol. 2011;25:897–907.
Myles C, Hodgson IA, Anthony N. Hollenberg. Prostate cancer cells is independent of NCoR and SMRT activity of androgen receptor antagonist bicalutamide in corepressors. Cancer Res. 2007;67:8388–95.
Amaral TM, Macedo D, Fernandes I, Costa L. Castration-resistant prostate cancer: mechanisms, targets, and treatment. Prostate Cancer. 2012;2012:327253.
Pienta KJ, Bradley D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin Cancer Res. 2006;12:1665–71.
Sen A, De Castro I, Defranco DB, Deng FM, Melamed J, Kapur P, et al. Paxillin mediates extranuclear and intranuclear signaling in prostate cancer proliferation. J Clin Invest. 2012;122:2469–81.
Zhu B, Kyprianou N. Transforming growth factor beta and prostate cancer. Cancer Treat Res. 2005;126:157–73.
Rojas A, Padidam M, Cress D, Grady WM. TGF-beta receptor levels regulate the specificity of signaling pathway activation and biological effects of TGF-beta. Biochim Biophys Acta. 2009;1793:1165–73.
Li Z, Habuchi T, Tsuchiya N, Mitsumori K, Wang L, Ohyama C, et al. Increased risk of prostate cancer and benign prostatic hyperplasia associated with transforming growth factor-beta 1 gene polymorphism at codon10. Carcinogenesis. 2004;25:237–40.
Wikstrom P, Damber J, Bergh A. Role of transforming growth factor-beta1 in prostate cancer. Microsc Res Tech. 2001;52:411–9.
Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett. 2012;586:1906–12.
Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-β targeted cancer therapy. Int J Biol Sci. 2012;8:964–78.
Heldin CH, Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res. 2012;347:21–36.
Bierie B, Moses HL. Transforming growth factor beta (TGF-beta) and inflammation in cancer. Cytokine Growth Factor Rev. 2010;21:49–59.
Itatani Y, Kawada K, Fujishita T, Kakizaki F, Hirai H, Matsumoto T, et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1(+) myeloid cells and facilitate liver metastasis. Gastroenterology. 2013;145:1064–75. e1011.
Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73:725–35.
Bacman D, Merkel S, Croner R, Papadopoulos T, Brueckl W, Dimmler A. TGF-beta receptor 2 downregulation in tumour-associated stroma worsens prognosis and high-grade tumours show more tumour-associated macrophages and lower TGF-beta1 expression in colon carcinoma: a retrospective study. BMC Cancer. 2007;7:156.
Dong M, How T, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, et al. The type III TGF-β receptor suppresses breast cancer progression. J Clin Invest. 2007;117:206–17.
Yang G, Yang X. Smad4-mediated TGF-β signaling in tumorigenesis. Int J Biol Sci. 2010;6:1–8.
Teixeira AL, Gomes M, Nogueira A, Azevedo AS, Assis J, Dias F, et al. Improvement of a predictive model of castration-resistant prostate cancer: functional genetic variants in TGFbeta1 signaling pathway modulation. PLoS One. 2013;8:e72419.
Kim SJ, Im YH, Markowitz SD, Bang YJ. Molecular mechanisms of inactivation of TGF-beta receptors during carcinogenesis. Cytokine Growth Factor Rev. 2000;11:159–68.
Wikstrom P, Stattin P, Franck-Lissbrant I, Damber JE, Bergh A. Transforming growth factor beta1 is associated with angiogenesis, metastasis, and poor clinical outcome in prostate cancer. Prostate. 1998;37:19–29.
Butz H, Racz K, Hunyady L, Patocs A. Crosstalk between TGF-beta signaling and the microRNA machinery. Trends Pharmacol Sci. 2012;33:382–93.
Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61.
Hata A, Davis BN. Control of microRNA biogenesis by TGFbeta signaling pathway-a novel role of Smads in the nucleus. Cytokine Growth Factor Rev. 2009;20:517–21.
Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–84.
Long X, Miano JM. Transforming growth factor-beta1 (TGF-beta1) utilizes distinct pathways for the transcriptional activation of microRNA 143/145 in human coronary artery smooth muscle cells. J Biol Chem. 2011;286:30119–29.
Ma S, Chan YP, Kwan PS, Lee TK, Yan M, Tang KH, et al. MicroRNA-616 induces androgen-independent growth of prostate cancer cells by suppressing expression of tissue factor pathway inhibitor TFPI-2. Cancer Res. 2011;71:583–92.
Shi XB, Tepper CG. deVere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–38.
Sk A. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors. 2012;12:3359–69.
Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3:311–30.
Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–95.
Teixeira AL, Ferreira M, Silva J, Gomes M, Dias F, Santos JI, Mauricio J, Lobo F, Medeiros R. Higher circulating expression levels of mir-221 associated with poor overall survival in renal cell carcinoma patients. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2013.
Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227.
Davis BN, Hata A. Regulation of microRNA biogenesis: a miRiad of mechanisms. Cell Commun Signal CCS. 2009;7:18.
Elston R, Inman GJ. Crosstalk between p53 and TGF-beta signalling. J Signal Transduct. 2012;2012:294097.
McDonald RA, Hata A, MacLean MR, Morrell NW, Baker AH. MicroRNA and vascular remodelling in acute vascular injury and pulmonary vascular remodelling. Cardiovasc Res. 2012;93:594–604.
Bowen T, Jenkins RH, Fraser DJ. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J Pathol. 2013;229:274–85.
Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–84.
Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, et al. TGF-beta activates akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–9.
Kato M, Wang L, Putta S, Wang M, Yuan H, Sun G, et al. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells. J Biol Chem. 2010;285:34004–15.
Wang B, Koh P, Winbanks C, Coughlan MT, McClelland A, Watson A, et al. miR-200a prevents renal fibrogenesis through repression of TGF-beta2 expression. Diabetes. 2011;60:280–7.
Dogar AM, Towbin H, Hall J. Suppression of latent transforming growth factor (TGF)-beta1 restores growth inhibitory TGF-beta signaling through microRNAs. J Biol Chem. 2011;286:16447–58.
Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907.
Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–9.
Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, Lagna G, et al. Down-regulation of Kruppel-like factor-4 (klf4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J Biol Chem. 2011;286:28097–110.
Jurkin J, Schichl YM, Koeffel R, Bauer T, Richter S, Konradi S, et al. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J Immunol. 2010;184:4955–65.
Bello-DeOcampo D, Tindall DJ. TGF-betal/Smad signaling in prostate cancer. Curr Drug Targets. 2003;4:197–207.
Paolo Fuzio PD. Monica Rutigliano, Michele Battaglia, et al. Regulation of TGF-b1 expression by androgen deprivation therapy of prostate cancer. Cancer Lett. 2012;318:135–44.
Zhang H, Cai X, Wang Y, Tang H, Tong D, Ji F. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep. 2010;24:1363–9.
Akao Y, Nakagawa Y, Iio A, Naoe T. Role of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia Jurkat cells. Leuk Res. 2009;33:1530–8.
Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10.
Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–92.
Ni Y, Meng L, Wang L, Dong W, Shen H, Wang G, et al. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene. 2013;517:197–204.
Fuse M, Nohata N, Kojima S, Sakamoto S, Chiyomaru T, Kawakami K, et al. Restoration of miR-145 expression suppresses cell proliferation, migration and invasion in prostate cancer by targeting FSCN1. Int J Oncol. 2011;38:1093–101.
Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, Barth S, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Molecular Cancer Res MCR. 2010;8:529–38.
Yin Y, Yan ZP, Lu NN, Xu Q, He J, Qian X, et al. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim Biophys Acta. 1829;2013:239–47.
Labbaye C, Testa U. The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J Hematol Oncol. 2012;5:13.
Lo U-G, Yang D, Hsieh J-T. The role of microRNAs in prostate cancer progression. Transl Androl Urol. 2013;2:228–41.
Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–24.
Xu B, Wang N, Wang X, Tong N, Shao N, Tao J, et al. Mir-146a suppresses tumor growth and progression by targeting EGFR pathway and in a p-ERK-dependent manner in castration-resistant prostate cancer. Prostate. 2012;72:1171–8.
Zhang J, Zhang D, Wu GQ, Feng ZY, Zhu SM. Propofol inhibits the adhesion of hepatocellular carcinoma cells by upregulating microRNA-199a and downregulating MMP-9 expression. Hepatobiliary Pancreat Dis Int. 2013;12:305–9.
Daneshmand S, Quek ML, Lin E, Lee C, Cote RJ, Hawes D, et al. Glucose-regulated protein GRP78 is up-regulated in prostate cancer and correlates with recurrence and survival. Hum Pathol. 2007;38:1547–52.
He J, Jing Y, Li W, Qian X, Xu Q, Li FS, et al. Roles and mechanism of miR-199a and miR-125b in tumor angiogenesis. PLoS One. 2013;8:e56647.
Landi L, Cappuzzo F. HER2 and lung cancer. Expert Rev Anticancer Ther. 2013;13:1219–28.
Carrion-Salip D, Panosa C, Menendez JA, Puig T, Oliveras G, Pandiella A, et al. Androgen-independent prostate cancer cells circumvent EGFR inhibition by overexpression of alternative her receptors and ligands. Int J Oncol. 2012;41:1128–38.
Baek KH, Hong ME, Jung YY, Lee CH, Lee TJ, Park ES, et al. Correlation of AR, EGFR, and HER2 expression levels in prostate cancer: immunohistochemical analysis and chromogenic in situ hybridization. Cancer Res Treat Off J Korean Cancer Assoc. 2012;44:50–6.
Wu D, Huang HJ, He CN, Wang KY. MicroRNA-199a-3p regulates endometrial cancer cell proliferation by targeting mammalian target of rapamycin (mTOR). Int J Gynecol Cancer Off J Int Gynecol Cancer Soc. 2013;23:1191–7.
Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. Mir-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–93.
Acknowledgments
We would like to thank the Liga Portuguesa Contra o Cancro-Centro Regional do Norte (Portuguese League Against Cancer) and FCT-Fundação para a Ciência e Tecnologia. ALT is a doctoral degree grant holder from FCT (SFRH/BD/47381/2008).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Juliana Inês Santos and Ana Luísa Teixeira contributed equally to this work.
Rights and permissions
About this article
Cite this article
Santos, J.I., Teixeira, A.L., Dias, F. et al. Restoring TGFβ1 pathway-related microRNAs: possible impact in metastatic prostate cancer development. Tumor Biol. 35, 6245–6253 (2014). https://doi.org/10.1007/s13277-014-1887-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-1887-z