Abstract
Land use management is a central challenge for the 21st century with unprecedented and competing demands to produce food, feed/fodder, fibre, fuel, and essential ecosystem services which sustain life. Global change requires rapid adaptation in current and emerging crops as well as in the foundation species of natural ecosystems. Revolutions in genomics and high throughput experimentation are transforming breeding so that adaptive traits in new environments can be predicted and selected more directly from germplasm collections of crops and wild species. This genomic breeding is now feasible in almost any species and has promise to help meet the need to feed and nourish over 9 billion people by 2050. Genomic techniques can accelerate our response to food security challenges of yield, quality and resilience and also address environmental security challenges. To achieve its potential there will need to be widespread and ongoing investments in the human capital to promote genomic breeding.
Similar content being viewed by others
Introduction
Food security is defined as having “physical and economic access to sufficient, safe and nutritious food, for an active and healthy life” (FAO 1996). To ensure continued food security, we must continue improving plant (i.e. crop) productivity, the basis of world agriculture, by exploiting the new genetic-sequencing and ‘big data’ technologies that are underpinning the ‘genomics revolution’ in life sciences (Varshney et al. 2014; Enriquez 1998). Application of advanced ‘genomic breeding’ technologies will accelerate crop improvement, and holds the promise of ‘personalised agriculture’ - crop species and varieties that are tailor-made for particular climates and environments. Such technologies will also allow plant breeders to effectively and simultaneously target new crop species and traits such as resilience, quality and yield which are crucial to food security. This plant breeding ‘revolution’ requires more human capital, ensuring sufficient and qualified plant breeders to carry out this work. It also needs further improvements in data collection, analysis, dissemination and re-usability.
Agriculture and the genomics revolution
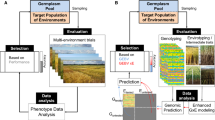
Breeding using genomic tools, or ‘genomic breeding’, has the potential to revolutionise plant breeding, providing farmers with more productive crops in a shorter amount of time. Plant breeders and scientists now utilise unprecedented amounts of information from high-throughput genetic sequencing and phenotyping technologies. Due to the low cost and high resolution, genomics-assisted breeding can move beyond traditional targets of narrow, single variable characteristics, toward breeding for whole-plant characteristics previously regarded as too technically-challenging (Zamir 2013). These approaches are analogous to the promise of ‘personalised medicine’ where treatment and lifestyle recommendations are tailored to the individual, based on his or her genetic sequence (Offit 2011). A parallel ‘personalised agriculture’ would look up which crop varieties and species are most-suitable to farmers for their specific current and future on-farm conditions (Furbank and Tester 2011).
The race is on to sequence the genomes of crop varieties and wild relatives (Zamir 2013; DivSeek 2014; specific examples: Krishnan et al. 2014; Lin et al. 2014) in an effort to describe the extent of genetic variation in current collections. With this information, powerful experiments can be designed to draw links between genomic sequence and crop performance and potential (Nordborg and Weigel 2008; Bergelson and Roux 2010; Myles et al. 2009).
So-called Genome Wide Association Studies (GWAS) comprehensively scan for genetic components that control heritable traits (Brachi et al. 2011; Myles et al. 2009; Nordborg and Weigel 2008). In GWAS, the historical relatedness of the individual lineages is first determined. Subsequently the causal trait loci are identified at high resolution such that markers tag specific genes, which control a particular trait. These so called Quantitative Trait Loci (QTL) may be quickly drawn from a wild relative into an elite line, bypassing tedious phenotypic screening. Genomic sequencing of baseline diversity populations are an essential prerequisite step. Techniques for gene and QTL discovery have been reviewed elsewhere (Varshney et al. 2014). These loci are genetic markers that can help predict traits via in-silico selection of breeding material, and accelerate the breeding process. We can predict, for example, the performance of a particular plant variety under various environmental conditions, without actually growing plants under those conditions, by examining the QTLs present in that variety’s genome. As this trait prediction improves, we can quickly recognize the value of new combinations of genetic variants without having to plant and measure each variety at each generation as is typical in traditional breeding.
GWAS and QTL mapping have already contributed substantially to crop improvement. QTL mapping studies have helped enrich vitamin A content of tomato (Ronen et al. 2000), sorghum (Fernandez et al. 2008) and melon (Cuevas et al. 2008), for example. In these studies, researchers applied QTL-mapping techniques to identify regions of the crop genomes that correlated with increased accumulation of specific carotenoid plant pigments, some of which are metabolic precursors of vitamin A.
Two factors, however, have substantially improved the usefulness of genomic breeding in recent years: next-generation sequencing (Smith et al. 2010) and high-throughput phenomics (Yang et al. 2013). The cost, in time and money, of genetic sequencing has dropped precipitously in the last few years through combined innovations in chemistry, robotics and computing (Smith et al. 2010; Harakalova et al. 2011). Genomic sequencing is faster, cheaper and more reliable than ever before. As a result, much more genetic data can be acquired by plant breeders and scientists, which means that the genetic make-up of individual lines and populations can be quickly assessed with near base pair resolution. Entire collections of key (crop) species are being sequenced. These can be exploited in several ways: one is to select powerful core sets of lines to be used for trait mapping across multiple farm locations and management practices (Brachi et al. 2011). In many cases, the output of these experiments, are becoming accessible to scientists everywhere (Schmitz and Zhang 2011; Wang et al. 2009; Varshney et al. 2014), but more widespread sharing of these kinds of data is necessary to accelerate progress.
Physical plant characteristics are also becoming easier to quantify. What would have once involved hours of labour by scientists armed with rulers and scales is now accomplished in minutes using robots and advanced imaging (Finkel 2009). This has meant greater volume and reliability of phenotypic data: measuring plant characteristics in the lab, greenhouse and even in the field has become far less subjective.
By exploiting these parallel technological advances in Genomics and Phenomics, plant breeding has the potential to become far more quantitative and predictive. The statistical power inherent in large coordinated sets of data allows antagonistic gene/QTL effects to be separated; an inability to do this has limited traditional breeding. It also means that multi-trait metrics of crop performance, for example resilience and nutritional quality, can be jointly selected rather than combining sequentially, saving several generations
Vast progress has also been made in techniques to analyse proteins and chemical compounds and the synergies between genomics (DNA), metabolomics (small compounds), transcriptomics (RNA), proteomics (proteins), and phenomics (physical characteristics) are expected to further enrich and accelerate plant breeding (Yang et al. 2013).
Genomic breeding for new (and old) target traits
Beyond producing sufficient quantities, or yield, farms will be expected to provide a stable supply of high-quality, nutritious food (Godfray et al. 2010). Reducing the environmental footprint and catering for local consumer preferences, are also worthy goals (Godfray et al. 2010; Swinton et al. 2007). Genomic breeding will be particularly useful for the introduction of wild traits into domesticated plants, and the improvement of orphan crops. In both cases, genomics and phenomics will accelerate breeding programs by helping predict plant performance reducing the need for onerous field studies. Table 1 shows a brief list of traits that can be targeted by genomic breeding for food and environmental security.
Quality
The nutritional quality of food is one of the pillars of food security (FAO 1996). It is thus sobering to note that malnutrition affects over 2 billion people worldwide (FAO 2013). Problems relating to poor diet are not limited to the poor: 20 % of the world’s population is obese (Obesity and overweight, WHO Factsheet 2014) and 80 % of all obesity-related deaths occur in developed countries. Pockets of malnutrition are scattered throughout even wealthy countries such as the United States (Obesity and overweight, WHO Factsheet 2014; Key Statistics and Graphics, Food and Nutrition Assistance, USDA 2014; Dubé et al. 2012)
An emphasis of plant breeding has been on yield for the global commodity market (feed, fuel, etc.) at the expense of nutrient quality or cultural preference such as fragrant rice (Bradbury et al. 2005). In the face of dietary problems and preferences, plant breeders and scientists are challenged to breed crops that are nutritious, which in some cases means reducing concentrations of saturated fats and easily-metabolised sugars. Several notable attempts have been made to enrich or bio-fortify crop plants.
One notable example of food quality-improvement is the breeding of high-fibre barley. Fibre is an essential component of the human diet, promoting beneficial intestinal bacteria (Tuohy et al. 2012). Preservation of such microbes has, in turn, been associated with decreased risk of bowel cancer and other health benefits (Kendall et al. 2010). The Commonwealth Scientific and Industrial Research Organisation in Australia (CSIRO) recently led a large barley breeding effort, which has resulted in a commercialised barley variety with roughly twice the fibre content of other barley varieties (The BARLEYmax Better Nutrition Report 2009).
This and other breeding efforts have demonstrated that crop nutritional quality is a multi-genic trait. Many genes and gene combinations that could have improved quality were lost during initial domestication (Xiao et al. 1996). This is because the genetic potential cannot always be seen in the phenotypes selected for in traditional breeding. Now with genomic breeding, coupled with high-throughput metabolomics for example, loci controlling specific metabolites such as essential oils can be screened and predicted in crop and wild populations (Henery et al. 2007; Fernandez et al. 2008). In conclusion, quality and quantity do not have to be a trade-off or optimized sequentially; both can be directly selected with genomic breeding.
Resilience
For better food security, crops of the future will also need to be more resilient to overcome the threats posed by changing and suboptimal growth conditions, such as resource limitation, and unfavourable weather. Past breeding efforts produced so-called ‘elite’ varieties, to be grown under ‘optimal’ conditions. However, farmers must create these desired environments through irrigation and fertiliser application, leaving a large environmental footprint. Not only will we need to reduce this footprint, but also most of the world’s food production, does not, and will not, take place under optimal conditions. Most farmers have only limited agronomic options.
Fertilisers and other inputs are limiting factors for food production in many developing countries; elite varieties thus almost always fail to reach their yield potential in many African, South American and Asian countries (Cassman 1999). Even in developed countries, volatility in fertiliser, chemical and fuel prices can limit crop inputs, with flow-on effects for production and food availability on world markets (Piesse and Thirtle 2009).
In addition, extreme weather events, including droughts, heatwaves, floods and heavy storms have increasingly contributed to dramatic food shortages in recent decades (Schmidhuber and Tubiello 2007). Every International Panel on Climate Change report, including the latest, warn that such events will be more frequent in the future, as the world’s climate changes (IPCC 2014). Many ‘resilience’ traits are being bred into crops: drought-resistant cereals which can survive water shortage, harsh heat, and light, and proceed to yield a crop, are very important for the future of Australian farming (Chaves et al. 2003). Flood-tolerant rice varieties are another example (Bailey-Serres et al. 2010).
Another way genomic breeding could contribute to resilient crops is through the increased potential to design generalist genotypes that can deal with a large variety of environments. In other words: instead of elite varieties that produce exceptional yields under ideal conditions, we could aim at breeding for resilient varieties that can deliver ‘good’ yields across a range of environmental conditions, spatial and temporal. Defining these resilience traits as compound phenotypes of sustainable yield allows their quantitative measurement and exploitation by genomic breeding. These resilient genotypes would contribute to food security in several ways: because they grow in a range of environments, they will be more widely available; because they are resilient to extreme weather events, they potentially make farming more predictable, which would not only stabilize the food supply but also the farmers’ income.
Although economic considerations have contributed to lack of interest in ‘generalist’ crops, these crops would also be difficult to breed from existing crop varieties given the likely loss of such traits during previous breeding efforts (Tanksley and McCouch 1997). Moreover, resilience traits are multi-genic, flood-tolerant rice being a notable exception (Septiningsih et al. 2009; Bailey-Serres et al. 2010; Xu et al. 2006). Thus, similar breeding considerations apply as described above for nutritional quality breeding: genomic breeding will also play a growing role in breeding for crop resilience.
Ecosystem services
Genomic breeding could also help improve food security by accelerating plant breeding for ecosystem services. Potentially, varieties from crop and non-crop species could be selected for the ecosystem services they provide: biodiversity-promotion (animals, plants and microbes), erosion control, water and soil health, climate change mitigation, etc. (Godfray and Garnett 2014; Sayer and Cassman 2013). Targeted breeding of crop and natural vegetation for ecosystem services may improve environmental security, both on-farm and on larger scales (Swinton et al. 2007).
Breeding for plants that promote soil biology and chemistry, for example (McSwiney et al. 2010), could become easier by applying genomic plant-breeding techniques alongside high-throughput microbiome sequencing (metagenomics) and non-invasive, below-ground plant analyses (Chaparro et al. 2012). Breeding towards perennial lifestyle is another strategy for improving soil health, and is already showing great promise for improving soil health on farms (Glover et al. 2010; Culman et al. 2013). Two strategies for generation of perennial crops have been proposed: direct domestication of perennial plants (T. S. Cox et al. 2006; DeHaan et al. 2005) and ‘wide hybridisation’ of existing crops with wild relatives. Genomic breeding would augment both these methods by helping to directly select useful genetic material in a single generation, a dramatic improvement for long lived species. QTLs that promote plant-microbe interactions and perenniality have, for example, been identified in backcross populations of Sorghum and their progenitor, ‘johnsongrass’ (Paterson et al. 1995; Cox et al. 2002).
Genomic breeding can also be applied to non-crop species to improve food and environmental security. Remediation of water tables via the planting of native tree species has been demonstrated (Eldridge and Freudenberger 2005): improvements in tree genomics may enable widespread breeding for this ‘ecosystem service’ (Neale and Kremer 2011). Detailed assessment of ecosystem services on a large scale is now possible through the use of cost-effective sensors and cameras. Environmental parameters can be measured and ecosystem development recorded through time lapse photography. These data can then serve as phenotypes for selection and breeding, greatly improving vegetation conservation efforts with benefits for environmental health and thus food security and society.
Orphan species
Breeding of non-mainstream crop (orphan) species will also be easier with genomic tools: we expect a surge in research on crop species that have received little or no attention so far. The number of crop species consumed globally over the past fifty years has narrowed (Khoury et al. 2014), which is the other side of the coin of the plant breeding success of the past. While food production has obviously increased overall, some have raised concerns that the world’s reliance on a smaller number of food crops may make global food security more susceptible to local interruptions in production (Alston et al. 2009; Ray et al. 2013). Moreover, for at least some populations, a shift from traditional to Western diets has decreased their nutritional intake (Graham et al. 2007; Kennedy et al. 2005; Ruel 2003).
Application of genomic tools has become very accessible (cheap and widespread), which should help expand breeding from main staples into orphan crops by removing economic barriers to their study (Varshney et al. 2012). A paucity of genomic resources has, for example, limited lentil breeding; a recent transcriptome sequencing effort will reverse this by providing molecular markers to assist breeding efforts (Sharpe et al. 2013). Traditional and/or regional crops that have been overlooked by past, commercial, breeding efforts will thus benefit from greater use of genomic breeding.
How to exploit the ‘omics revolution’ for better food security
So, what needs to happen for agriculture and the world to get the most out of genomic breeding?
More hands on deck, for a start. The world desperately needs more plant breeders and scientists (Jones 2014). Over the last three decades, and despite phenomenal advances in life sciences, plant science has failed to capture the imagination of this generation’s (young) scientists. Whilst the number of PhD students enrolled in biomedical research doubled between 1982 and 2012, plant science PhD enrolments flat-lined (Alberts et al. 2014; Jones 2014). This is at least in part due to the much-lamented stagnation in worldwide agricultural and plant science research funding (Butler 2008). It is also likely due to a perception, in the developed world, that the food security struggle is over. Researchers, industry and governments must work together to remind the public that food security cannot be taken for granted, and to inspire more of our brightest young people to pursue careers in agricultural and plant sciences.
Not only do we need more of them, tomorrow’s plant scientists should also be data-savvy: the high-throughput approaches, and all –omics disciplines, are already producing data amounts that must be processed by computers. The immediate challenge to integrate them all will require sophisticated approaches and the ability to communicate across disciplines. Non-life science technologies, such as remote and automated environmental analytic technologies by satellites, drones and miniaturised chemical analysers (ConservationDrones.org 2014; Murphy et al. 2009) will make important contributions.
Despite the flood of plant data already available, there is room for more, and better, data collection, helping researchers to work more closely with breeders, farmers and landholders. Increasing the quality and availability of crop phenotypic data will increase the potential of genomic informed breeding to improve crops.
New, global scientific standards for biological data archiving and sharing are needed (Zamir 2013); there must also be incentives to contribute. Large amounts of phenotypic data are being collected each year as part of plant breeding research. These data are frequently unavailable to the broader scientific community, either because researchers are unwilling to share or do not have the means to do so, with the consequence of unnecessary duplication, and, more significantly, missed opportunities. Even when such data are available, they are often of limited use: disparate data formats and lack of detail regarding how data were obtained (meta-data) mean it is often impractical to analyse phenotypic data collected by others (Walls et al. 2012). Zamir has suggested reforms be made to funding conditions and data standards to allow greater retention, curation and distribution of such data, in the same way much data from more-sophisticated biochemical and genetic experiments are already available (Zamir 2013, 2014).
Plant research initiatives in many countries and regions are conducting trials of crop varieties across various climates and environments. High-throughput approaches could allow us to increase the scale such that most agricultural regions in the world would contribute data on crop varietal performance. The performance of genotypes in real-life environments under real life farming practices would provide a level of replication that allows robust inference about genotype-environment-management interactions. This will vastly increase the ability to tailor genotypes and, for example, provide better-suited crops for suboptimal environments, potentially benefitting farmers in developing countries.
Conclusion
Advanced plant science and genomics have revolutionised breeding and crop improvement, and will continue to do so. Innovation in collecting genotypes, phenotypes, and intermediate characteristics, is allowing new crop varieties to be selected faster and more accurately than ever before. With genomic techniques researchers can help address food security challenges of yield, quality, resilience, and other environmental and social needs. Investing in the human capital to perform genomic breeding is needed to improve food security, environments and livelihoods.
References
Alberts, B., Kirschner, M. W., Tilghman, S., & Varmus, H. (2014). Rescuing US biomedical research from its systemic flaws. Proceedings of the National Academy of Sciences, 111(16), 5773–5777. doi:10.1073/pnas.1404402111.
Alston, J. M., Beddow, J. M., & Pardey, P. G. (2009). Agricultural research, productivity, and food prices in the long run. Science, 325(5945), 1209–1210. doi:10.1126/science.1170451.
Bailey-Serres, J., Fukao, T., Ronald, P., Ismail, A., Heuer, S., & Mackill, D. (2010). Submergence tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice, 3(2–3), 138–147. doi:10.1007/s12284-010-9048-5.
Bergelson, J., & Roux, F. (2010). Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. [10.1038/nrg2896]. Nature Reviews Genetics, 11(12), 867–879.
Brachi, B., Morris, G., & Borevitz, J. (2011). Genome-wide association studies in plants: the missing heritability is in the field. Genome Biology, 12(10), 232.
Bradbury, L. M. T., Fitzgerald, T. L., Henry, R. J., Jin, Q., & Waters, D. L. E. (2005). The gene for fragrance in rice. Plant Biotechnology Journal, 3(3), 363–370. doi:10.1111/j.1467-7652.2005.00131.x.
Butler, D. (2008). Food crisis spurs research spending. http://www.nature.com/news/2008/080430/full/453008a.html. Accessed 27 October 2014 2008.
Cassman, K. G. (1999). Ecological intensification of cereal production systems: yield potential, soil quality, and precision agriculture. Proceedings of the National Academy of Sciences, 96(11), 5952–5959. doi:10.1073/pnas.96.11.5952.
Chaparro, J., Sheflin, A., Manter, D., & Vivanco, J. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biology and Fertility of Soils, 48(5), 489–499. doi:10.1007/s00374-012-0691-4.
Chaves, M. M., Maroco, J., #227, P., o., Pereira, J., & S., o. (2003). Understanding plant responses to drought - from genes to the whole plant. Functional Plant Biology, 30(3), 239-264, doi:http://dx.doi.org/10.1071/FP02076.
ConservationDrones.org (2014). http://conservationdrones.org/. Accessed 6 November 2014.
Cox, C. M., Murray, T. D., & Jones, S. S. (2002). Perennial wheat germ plasm lines resistant to eyespot, cephalosporium stripe, and wheat streak mosaic. Plant Disease, 86(9), 1043–1048. doi:10.1094/pdis.2002.86.9.1043.
Cox, T. S., Glover, J. D., Van Tassel, D. L., Cox, C. M., & DeHaan, L. R. (2006). Prospects for developing perennial grain crops. BioScience, 56(8), 649–659. doi:10.1641/0006-3568(2006)56[649:pfdpgc]2.0.co;2.
Cuevas, H. E., Staub, J. E., Simon, P. W., Zalapa, J. E., & McCreight, J. D. (2008). Mapping of genetic loci that regulate quantity of beta-carotene in fruit of US Western Shipping melon (Cucumis melo L.). Theoretical and Applied Genetics, 117(8), 1345–1359. doi:10.1007/s00122-008-0868-2.
Culman, S. W., Snapp, S. S., Ollenburger, M., Basso, B., & DeHaan, L. R. (2013). Soil and water quality rapidly responds to the perennial grain kernza wheatgrass. Agronomy Journal, 105(3), 735–744. doi:10.2134/agronj2012.0273.
DeHaan, L. R., Van Tassel, D. L., & Cox, T. S. (2005). Perennial grain crops: A synthesis of ecology and plant breeding. Renewable Agriculture and Food Systems, 20(01), 5–14. doi:10.1079/RAF200496.
DivSeek (2014). http://www.divseek.org/. Accessed 14 November 2014.
Dubé, L., Pingali, P., & Webb, P. (2012). Paths of convergence for agriculture, health, and wealth. Proceedings of the National Academy of Sciences, 109(31), 12294–12301. doi:10.1073/pnas.0912951109.
Eldridge, D. J., & Freudenberger, D. (2005). Ecosystem wicks: woodland trees enhance water infiltration in a fragmented agricultural landscape in eastern Australia. Austral Ecology, 30(3), 336–347. doi:10.1111/j.1442-9993.2005.01478.x.
Enriquez, J. (1998). Genomics and the world’s economy. Science, 281(5379), 925–926. doi:10.1126/science.281.5379.925.
FAO (1996). Rome declaration on world food security. Rome, Italy.
FAO (2013). State of food insecurity in the world. Rome: Food and Agriculture Organisation of the United Nations (FAO).
Fernandez, M. G. S., Hamblin, M. T., Li, L., Rooney, W. L., Tuinstra, M. R., & Kresovich, S. (2008). Quantitative trait loci analysis of endosperm color and carotenoid content in sorghum grain. Crop Science, 48(5), 1732–1743. doi:10.2135/cropsci2007.12.0684.
Finkel, E. (2009). With ‘phenomics’, plant scientists hope to shift breeding into overdrive. Science, 325(5939), 380–381. doi:10.1126/science.325_380.
Furbank, R. T., & Tester, M. (2011). Phenomics – technologies to relieve the phenotyping bottleneck. Trends in Plant Science, 16(12), 635–644. doi:10.1016/j.tplants.2011.09.005.
Glover, J. D., Reganold, J. P., Bell, L. W., Borevitz, J., Brummer, E. C., Buckler, E. S., et al. (2010). Increased food and ecosystem security via perennial grains. Science, 328(5986), 1638–1639. doi:10.1126/science.1188761.
Godfray, H. C. J., & Garnett, T. (2014). Food security and sustainable intensification. Philosophical Transactions of the Royal Society B: Biological Sciences, 369(1639), doi:10.1098/rstb.2012.0273.
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science, 327(5967), 812–818. doi:10.1126/science.1185383.
Graham, R. D., Welch, R. M., Saunders, D. A., Ortiz-Monasterio, I., Bouis, H. E., Bonierbale, M., et al. (2007). Nutritious subsistence food systems. In L. S. Donald (Ed.), Advances in Agronomy (Vol. Volume 92, pp. 1-74): Academic Press.
Key Statistics and Graphics, Food and Nutrition Assistance, USDA (2014). http://www.ers.usda.gov/topics/food-nutrition-assistance/food-security-in-the-us/key-statistics-graphics.aspx#.VEiKafmUeCk. Accessed 23 October 2014 2014.
Harakalova, M., Mokry, M., Hrdlickova, B., Renkens, I., Duran, K., & van Roekel, H. (2011). Multiplexed array-based and in-solution genomic enrichment for flexible and cost-effective targeted next-generation sequencing. Natural Protocols, 6(12), 1870–1886. doi:10.1038/nprot.2011.396. http://www.nature.com/nprot/journal/v6/n12/abs/nprot.2011.396.html#supplementary-information.
Henery, M. L., Moran, G. F., Wallis, I. R., & Foley, W. J. (2007). Identification of quantitative trait loci influencing foliar concentrations of terpenes and formylated phloroglucinol compounds in Eucalyptus nitens. New Phytologist, 176(1), 82–95. doi:10.1111/j.1469-8137.2007.02159.x.
IPCC (2014). Climate Change 2014: Impacts, adaptation and vulnerablity, . Contribution of Working Group II to the Fifth Assessment Report of the Intergovenrmental Panel on Climate Change.
Jones, A. M. (2014). Opinion: The planet needs more plant scientists. http://www.the-scientist.com/?articles.view/articleNo/41133/title/Opinion--The-Planet-Needs-More-Plant-Scientists/. Accessed 6 November 2014.
Kendall, C. W. C., Esfahani, A., & Jenkins, D. J. A. (2010). The link between dietary fibre and human health. Food Hydrocolloids, 24(1), 42–48. doi:10.1016/j.foodhyd.2009.08.002.
Kennedy, G., Islam, O., Eyzaguirre, P., & Kennedy, S. (2005). Field testing of plant genetic diversity indicators for nutrition surveys: rice-based diet of rural Bangladesh as a model. Journal of Food Composition and Analysis, 18(4), 255–268. doi:10.1016/j.jfca.2004.10.002.
Khoury, C. K., Bjorkman, A. D., Dempewolf, H., Ramirez-Villegas, J., Guarino, L., Jarvis, A., et al. (2014). Increasing homogeneity in global food supplies and the implications for food security. Proceedings of the National Academy of Sciences, 111(11), 4001–4006. doi:10.1073/pnas.1313490111.
Krishnan, S., Waters, G. D. L. E., & Henry, R. J. (2014). Australian wild rice reveals pre-domestication origin of polymorphism deserts in rice genome. PLoS ONE, 9(6), e98843. doi:10.1371/journal.pone.0098843.
Lin, T., Zhu, G., Zhang, J., Xu, X., Yu, Q., & Zheng, Z. (2014). Genomic analyses provide insights into the history of tomato breeding. Nature Genetics, 46(11), 1220–1226. doi:10.1038/ng.3117. http://www.nature.com/ng/journal/v46/n11/abs/ng.3117.html#supplementary-information.
McSwiney, C. P., Snapp, S. S., & Gentry, L. E. (2010). Use of N immobilization to tighten the N cycle in conventional agroecosystems. Ecological Applications, 20(3), 648–662. doi:10.1890/09-0077.1.
Murphy, D. V., Osman, M., Russell, C. A., Darmawanto, S., & Hoyle, F. C. (2009). Potentially mineralisable nitrogen: relationship to crop production and spatial mapping using infrared reflectance spectroscopy. Soil Research, 47(7), 737–741. doi:10.1071/SR08096.
Myles, S., Peiffer, J., Brown, P. J., Ersoz, E. S., Zhang, Z., Costich, D. E., et al. (2009). Association mapping: critical considerations shift from genotyping to experimental design. The Plant Cell, 21(8), 2194–2202.
Neale, D. B., & Kremer, A. (2011). Forest tree genomics: growing resources and applications. [10.1038/nrg2931]. Nature Reviews Genetics, 12(2), 111–122.
Nordborg, M., & Weigel, D. (2008). Next-generation genetics in plants. Nature, 456(7223), 720–723. doi:10.1038/nature07629.
Obesity and overweight, WHO Factsheet (2014). http://www.who.int/mediacentre/factsheets/fs311/en/. Accessed 23 October 2014 2014.
Offit, K. (2011). Personalized medicine: new genomics, old lessons. Human Genetics, 130(1), 3–14. doi:10.1007/s00439-011-1028-3.
Paterson, A. H., Schertz, K. F., Lin, Y. R., Liu, S. C., & Chang, Y. L. (1995). The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proceedings of the National Academy of Sciences, 92(13), 6127–6131.
Piesse, J., & Thirtle, C. (2009). Three bubbles and a panic: an explanatory review of recent food commodity price events. Food Policy, 34(2), 119–129. doi:10.1016/j.foodpol.2009.01.001.
Ray, D. K., Mueller, N. D., West, P. C., & Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS ONE, 8(6), e66428. doi:10.1371/journal.pone.0066428.
Ronen, G., Carmel-Goren, L., Zamir, D., & Hirschberg, J. (2000). An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proceedings of the National Academy of Sciences, 97(20), 11102–11107. doi:10.1073/pnas.190177497.
Ruel, M. T. (2003). Operationalizing dietary diversity: a review of measurement issues and research priorities. The Journal of Nutrition, 133(11), 3911S–3926S.
Sayer, J., & Cassman, K. G. (2013). Agricultural innovation to protect the environment. Proceedings of the National Academy of Sciences, 110(21), 8345–8348. doi:10.1073/pnas.1208054110.
Schmidhuber, J., & Tubiello, F. N. (2007). Global food security under climate change. Proceedings of the National Academy of Sciences, 104(50), 19703–19708. doi:10.1073/pnas.0701976104.
Schmitz, R. J., & Zhang, X. (2011). High-throughput approaches for plant epigenomic studies. Current Opinion in Plant Biology, 14(2), 130–136. doi:10.1016/j.pbi.2011.03.010.
Septiningsih, E. M., Pamplona, A. M., Sanchez, D. L., Neeraja, C. N., Vergara, G. V., Heuer, S., et al. (2009). Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Annals of Botany, 103(2), 151–160. doi:10.1093/aob/mcn206.
Sharpe, A., Ramsay, L., Sanderson, L.-A., Fedoruk, M., Clarke, W., Li, R., et al. (2013). Ancient orphan crop joins modern era: gene-based SNP discovery and mapping in lentil. BMC Genomics, 14(1), 192.
Smith, A. M., Heisler, L. E., St.Onge, R. P., Farias-Hesson, E., Wallace, I. M., & Bodeau, J. (2010). Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Research, 38(13), e142. doi:10.1093/nar/gkq368.
Swinton, S. M., Lupi, F., Robertson, G. P., & Hamilton, S. K. (2007). Ecosystem services and agriculture: cultivating agricultural ecosystems for diverse benefits. Ecological Economics, 64(2), 245–252. doi:10.1016/j.ecolecon.2007.09.020.
Tanksley, S. D., & McCouch, S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science, 277(5329), 1063–1066. doi:10.1126/science.277.5329.1063.
The BARLEYmax Better Nutrition Report (2009). http://www.csiro.au/Outcomes/Food-and-Agriculture/BARLEYmax_FFF_Report.aspx. Accessed 6 November 2014.
Tuohy, K. M., Conterno, L., Gasperotti, M., & Viola, R. (2012). Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. Journal of Agricultural and Food Chemistry, 60(36), 8776–8782. doi:10.1021/jf2053959.
Varshney, R. K., Ribaut, J.-M., Buckler, E. S., Tuberosa, R., Rafalski, J. A., & Langridge, P. (2012). Can genomics boost productivity of orphan crops? [Opinion and Comment]. Natural Biotechnology, 30(12), 1172–1176. doi:10.1038/nbt.2440. http://www.nature.com/nbt/journal/v30/n12/abs/nbt.2440.html#supplementary-information.
Varshney, R. K., Terauchi, R., & McCouch, S. R. (2014). Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biology, 12(6), e1001883. doi:10.1371/journal.pbio.1001883.
Walls, R. L., Athreya, B., Cooper, L., Elser, J., Gandolfo, M. A., Jaiswal, P., et al. (2012). Ontologies as integrative tools for plant science. American Journal of Botany, 99(8), 1263–1275. doi:10.3732/ajb.1200222.
Wang, Z., Gerstein, M., & Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics, 10(1), 57–63. doi:10.1038/nrg2484.
Xiao, J., Grandillo, S., Ahn, S. N., McCouch, S. R., Tanksley, S. D., Li, J., et al. (1996). Genes from wild rice improve yield. Nature, 384(6606), 223–224. doi:10.1038/384223a0.
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., & Heuer, S. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature, 442(7103), 705–708. doi:10.1038/nature04920. http://www.nature.com/nature/journal/v442/n7103/suppinfo/nature04920_S1.html.
Yang, W., Duan, L., Chen, G., Xiong, L., & Liu, Q. (2013). Plant phenomics and high-throughput phenotyping: accelerating rice functional genomics using multidisciplinary technologies. Current Opinion in Plant Biology, 16(2), 180–187. doi:10.1016/j.pbi.2013.03.005.
Zamir, D. (2013). Where have all the crop phenotypes gone? PLoS Biology, 11(6), e1001595. doi:10.1371/journal.pbio.1001595.
Zamir, D. (2014). A wake-up call with coffee. Science, 345(6201), 1124. doi:10.1126/science.1258941.
Acknowledgments
This paper was part of a workshop sponsored by the OECD Co-operative Research Programme on Biological Resource Management for Sustainable Agricultural Systems.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Rivers, J., Warthmann, N., Pogson, B.J. et al. Genomic breeding for food, environment and livelihoods. Food Sec. 7, 375–382 (2015). https://doi.org/10.1007/s12571-015-0431-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12571-015-0431-3