Abstract
The rising and setting of the sun marks a transition between starkly contrasting environmental conditions for vegetative life. Given these differing diurnal and nocturnal environmental factors and the inherent regularity of the transition between the two, it is perhaps unsurprising that plants have developed an internal timing mechanism (known as a circadian clock) to allow modulation of gene expression and metabolism in response to external cues. Entrainment of the circadian clock, primarily via the detection of changes in light and temperature, maintains synchronization between the surrounding environment and the endogenous clock mechanism. In this review, recent advances in our understanding of the molecular workings of the plant circadian clock are discussed as are the input pathways necessary for entrainment of the clock machinery.
Similar content being viewed by others
The Importance of Circadian Clocks
The rotation of the Earth acts to confer regular environmental changes in light availability and temperature and plants alter their physiology, biochemistry, and metabolism in response to these abiotic cues over the course of a day (Hall and McWatters 2006). In addition, the inherent regularity of the transition between day and night also allows alterations in temperature and light to be predictive of subsequent abiotic stresses. For example, dusk is typically accompanied by a decrease in temperature and possible frost. It is therefore unsurprising that plants have developed an internal timing mechanism (referred to as a circadian clock) that allows prescient alterations in gene expression and biochemistry. Indeed, the circadian clock causes the regular oscillation of between 30% and 40% of genes in the model plant Arabidopsis thaliana (Arabidopsis), even when grown under constant light and temperature (Covington et al. 2008). These broad changes in gene expression precipitate a range of physiological responses including the regulation of hypocotyl growth (Dowson-Day and Millar 1999; Nozue et al. 2007), alterations to plant hormone production and sensitivity (Covington and Harmer 2007; Michael et al. 2008a, b; Robertson et al. 2009) and time of flowering (Imaizumi and Kay 2006). In concert, such changes promote the fitness of plants grown in synchrony between endogenous and environmental cues (Green et al. 2002; Dodd et al. 2005b).
Circadian clocks are conceptually thought to be comprised of three parts: a central oscillator typically consisting of a negative feedback loop, input pathways to allow entrainment to local environmental conditions, and output pathways which act to modulate responses dependent on these endogenous cues. Although at its most basic level a circadian clock can consist of a single negative feedback loop with input and output pathways (Dong and Golden 2008), evolution has typically led to the development of multiple interconnected molecular oscillators with varied levels of redundancy (Dardente and Cermakian 2007; Dunlap et al. 2007; Dubruille and Emery 2008; Harmer 2009). The inclusion of partially redundant interlocking components likely allows greater flexibility in the modulation of clock inputs during evolution (such as by altering sensitivity to light and temperature in different climates; Michael et al. 2003a) while also allowing greater accuracy of the timing mechanism itself (Rand et al. 2004; Stelling et al. 2004). In addition to these core concepts, circadian clocks are recognised as having additional properties including temperature compensation and “gating”. Temperature compensation allows a circadian clock to oscillate at approximately the same frequency over a broad range of physiological temperatures (from 12°C to 27°C; Edwards et al. 2005) while gating refers to the regulated sensitivity of the central oscillator to input stimuli. This latter mechanism enables the circadian clock to persist in plants grown in constant experimental conditions by reducing the responsiveness of core components to light during the subjective night (Harmer 2000; Covington et al. 2001; Carre 2002).
The Arabidopsis Circadian Clock
Although circadian clocks are found across both prokaryotic and eukaryotic phyla, the components that comprise these internal oscillators are not conserved (Dunlap et al. 1999; Hardin 2005; Brunner and Schafmeier 2006; Harmer 2009). Instead, it appears that clocks have arisen independently in multiple lineages. Much of the work studying the plant circadian clock has been completed using Arabidopsis in combination with bioluminescent reporter constructs which provide a visual output of the endogenous circadian rhythm (Millar et al. 1992), although leaf movement and qPCR have also been used to monitor clock activity (McClung 2006).
Using these techniques, mutant Arabidopsis have been isolated that differ in period (time from one peak of expression to the next; Fig. 1), phase (relative time of day a peak of gene expression occurs), and peak amplitude (degree of oscillation from the median). Analysis of these mutants has revealed that the Arabidopsis central oscillator consists of at least three interlocking loops of gene expression in which transcription factors promote the transcription of their own negative regulators (Fig. 2). The first transcriptional negative feedback loop identified in Arabidopsis includes two related MyB-like transcription factors (CIRCADIAN CLOCK ASSOCIATED 1; CCA1 and LATE ELONGATED HYPOCOTYL; LHY) and a gene of unknown biochemical function known as TIMING OF CAB EXPRESSION 1 (TOC1; Alabadi et al. 2001). TOC1 promotes expression of CCA1 and LHY by an indirect mechanism (via factor X) while CCA1 and LHY bind directly to the promoter of TOC1 and repress its activity (Alabadi et al. 2001). Although this model has provided an invaluable basis for our understanding of the Arabidopsis clock, it is now apparent that this one-loop model is insufficient to explain all the available experimental data. For example, CCA1 HIKING EXPEDITION (CHE) has recently been shown to bind to the CCA1 promoter and to repress its transcriptional activity, possibly by interfering with TOC1 activity (Pruneda-Paz et al. 2009). Additionally, mathematical modelling techniques have suggested the existence of at least two other transcriptional feedback loops that interlock with that formed by CCA1/LHY/TOC1 (Locke et al. 2006; Zeilinger et al. 2006). The first of these additional loops introduces an unknown factor (Y; Fig. 2), which positively regulates TOC1 expression while being repressed itself by CCA1, LHY, and TOC1. Although there are indications that the gene GIGANTEA (GI) fulfils some of the requirements of factor Y (Locke et al. 2005, 2006), it is possible that GI instead acts to regulate TOC1 solely at a posttranscriptional level (Kim et al. 2007; Martin-Tryon et al. 2007; Sawa et al. 2007).
An idealized example of a circadian-regulated gene. During 2 days of entrainment to a 24-h photoperiod expression of this gene peaks in the evening with the period and phase of this expression synchronized to the alternating light conditions. Upon transfer to constant light, rhythmic gene expression continues due to the endogenous clock mechanism. Zeitgeber time indicates time elapsed since the start of each photoperiod. Gray boxes represent subjective night
The Arabidopsis circadian clock consists of multiple loops. Transcriptional feedback loops act at the core of the oscillator with regulation of protein stability adding robustness to this rhythm. In the morning-phased loop of the clock CCA1 and LHY negatively regulate TOC1 expression while promoting expression of PRR7 and PRR9. In the evening, TOC1 acts to promote expression of CCA1 and LHY, at least in part via an indirect mechanism involving repression of CHE activity. PRR5 is thought to repress expression of PRR7 and PRR9. PRR3, GI, and ZTL act in concert to regulate TOC1 and PRR5 protein stability in a light-dependent fashion. X and Y show as yet uncharacterized components as suggested by mathematical modeling. Genes (rectangles) and protein products (ovals) thought to act primarily in the morning are colored blue; evening-phased loops are shown in red. Solid gray lines show transcriptional regulation while bold solid lines detail proposed protein–protein interactions. Dashed lines are used to indicate transcription and translation
The third loop proposed to form a component of the Arabidopsis core oscillator involves proteins related to TOC1. These PSUEDO-RESPONSE REGULATOR genes (PRR3, PRR5, PRR7, and PRR9) share a common protein domain structure to TOC1 and are similarly expressed with a circadian rhythm, although the phase of peak expression of each of these genes is shifted by 2–3 h (with sequential peaks of expression from dawn of PRR9, PRR7, PRR5, PRR3, and TOC1, respectively; Matsushika et al. 2000). Although single mutational lesions in each PRR gene induce mild circadian rhythm defects (Eriksson et al. 2003; Farre et al. 2005; Para et al. 2007), PRR proteins have partial functional redundancy as higher order prr mutants display more pronounced circadian phenotypes (Farre et al. 2005; Nakamichi et al. 2005a, b, 2007, 2009; Salome and McClung 2005a). For instance, the prr5 prr7 prr9 triple mutant is essentially arrhythmic under all entrainment conditions tested (Nakamichi et al. 2007). Through a combination of experimental and computational methods, it has been proposed that CCA1 and LHY act to promote PRR7 and PRR9 expression (Farre et al. 2005; Locke et al. 2006; Zeilinger et al. 2006) while PRR7 overexpression has been shown to reduce CCA1 and LHY mRNA levels (Farre and Kay 2007), thereby forming an additional morning-phased loop within the central oscillator (Fig. 2). Overexpression of PRR5 causes repression of PRR7 and PRR9 (Sato et al. 2002), suggesting an antagonistic role for PRR5 in the regulation of morning-phased genes.
The Effect of Light on the Circadian Clock
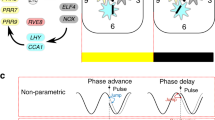
While oscillations in gene expression derived from the Arabidopsis circadian clock can be detected in seedlings germinated in the absence of environmental cues beyond inbibition of seed (Zhong et al. 1998; Salome et al. 2008), the circadian circuitry is capable of being entrained by stimuli such as light and temperature (Fankhauser and Staiger 2002; Cashmore 2003; Panda et al. 2003; Millar 2004). Such exogenous cues are often referred to as “zeitgebers” (or “time-givers”) and entrainment by these signals allows resetting of the clock to local conditions such as day length. The clock mechanism displays differential sensitivity to zeitgebers over the course of a day. For example, light pulses applied in the early morning cause an advancement of the clock mechanism whereas evening pulses delay the oscillator (Devlin and Kay 2001). This time-dependent effect causes phase advancement in the morning and phase delay in the late afternoon under normal diurnal conditions, allowing constant readjustment of the clock to prevailing conditions (Devlin and Kay 2001).
In order to detect light, plants express a suite of photoreceptors that confer sensitivity across the visual spectrum. The red portion of the spectrum is primarily sensed by phytochromes (Rockwell et al. 2006) while cryptochromes, phototropins, and zeitlupe family proteins are sensitive to blue light (Briggs 2007; Christie 2007; Kim et al. 2007; Li and Yang 2007). The latter family consists of three members (ZEITLUPE/ZTL, FLAVIN-BINDING KELCH DOMAIN F BOX PROTEIN1/FKF1, and LOV KELCH PROTEIN2) of which ZTL and FKF1 are known to have light-dependent functionality (Kim et al. 2007; Sawa et al. 2007). Light regulation of the circadian clock occurs within multiple loops of the circadian clock at both transcriptional, posttranscriptional, and posttranslational levels (Gutierrez et al. 2002; Lidder et al. 2005; Kim et al. 2007; Yakir et al. 2007), with phytochromes, cryptochromes, and zeitlupe family proteins playing primary roles in this regulation (Somers et al. 1998a, 2000; Devlin and Kay 2000; Schultz et al. 2001; Kim et al. 2007). Phytochrome and cryptochrome mutants have longer periods than wild-type only when monitored under constantly lit conditions (Somers et al. 1998a; Devlin and Kay 2000), suggesting that they act within the light input component of the clock rather than the central oscillator itself. In contrast, ztl mutants have a long period when maintained in either constant light or darkness (Somers et al. 2004), suggesting a more integral role within the central clock apparatus for this latter gene. Phototropin1 mutants do not display any circadian defects (Devlin and Kay 2001), suggesting that phototropins do not have a role in light input into the circadian clock (Salome and McClung 2005b).
Although it is clear that phytochromes and cryptochromes have key roles in coordinating and maintaining circadian rhythms, the identity and function of intermediate factors linking these photoreceptors with core circadian components remain obscure. Good candidates for intermediate components include the similarly named but unrelated genes EARLY FLOWERING 3 and 4 (ELF3 and ELF4) which are necessary for the gating of light signals input into the clock (McWatters et al. 2000, 2007; Covington et al. 2001). XAP5 CIRCADIAN TIMEKEEPER (Martin-Tryon and Harmer 2008) and SENSITIVITY TO RED LIGHT REDUCED (Staiger et al. 2003) also have circadian and light signaling defects, suggesting that they may act in both pathways. What is clear, however, is that light inputs feed into the circadian clock at multiple points in several clock loops. For instance, the expression levels of many clock genes (including CCA1, LHY, PRR7, PRR9, and GIGANTEA; GI) are upregulated by light (Wang and Tobin 1998; Martinez-Garcia et al. 2000; Eriksson et al. 2003; Farre et al. 2005; Locke et al. 2005; Farre and Kay 2007). Indeed, mutations in PRR7 and PRR9 only cause a long period in constantly lit conditions (Eriksson et al. 2003; Farre et al. 2005) suggesting that these genes act in pathways to modulate the effect of light on the entrainment of the circadian clock.
Although little is known regarding the biochemical function of TOC1, recent years have seen steady gains in our knowledge of light-mediated protein stability of this protein. Work utilizing a tagged TOC1 transgene initially revealed that TOC1 protein levels are stabilized by light (Mas et al. 2003) and subsequent analysis has identified several key components of this regulatory pathway. TOC1 is degraded in darkness by a SKP/CULLIN/F-box ubiquitin ligase complex containing CULLIN1 and ZTL, which contains an F-box domain (SCFZTL; Mas et al. 2003; Harmon et al. 2008). The ability of SCFZTL to bind and degrade TOC1 is regulated by PRR3 and GI as well as by light; a light-dependent binding between ZTL and GI prevents degradation of either protein while a physical interaction between PRR3 and TOC1 stymies recruitment and subsequent degradation of TOC1 by SCFZTL (Kim et al. 2007; Para et al. 2007; Fujiwara et al. 2008). It is attractive to suppose that upon nightfall ZTL, newly released from the light-requiring ZTL/GI complex binds TOC1 and causes its degradation (Somers et al. 2007). The accumulation of inactive ZTL over the course of a day enables a rapid decrease in TOC1 protein levels upon the onset of night, thereby enhancing the robustness of the transcriptional feedback loops of which TOC1 is a part (Fig. 2; Somers et al. 2007). In a similar manner, PRR5 protein accumulates in the evening before being targeted for degradation by ZTL (Kiba et al. 2007; Fujiwara et al. 2008), leading to the suggestion that PRR5, TOC1, GI, and ZTL act as a functional unit within the evening-phased loop of the Arabidopsis clock (Fujiwara et al. 2008).
Clock Entrainment by Temperature
The other dominant zeitgeber of the Arabidopsis circadian clock is temperature, with steps as small as 4°C being capable of entraining the clock mechanism (Somers et al. 1998b; Salome and McClung 2005a). The majority of large-scale mutant screens to identify Arabidopsis clock genes have used light as an entrainment signal and it is therefore unsurprising that comparatively little is known about temperature-sensitive entrainment of the clock. It does appear, however, that the circadian regulation of individual genes may differ based upon the entrainment conditions used; CAB2 and TOC1 expression are similarly modulated by light and temperature whereas the phase of CAT3 expression is more sensitive to changes in temperature than light (Michael et al. 2003b). Temperature inputs into the clock are at least in part incorporated via loops containing PRR7 and PRR9 as a prr7 prr9 double mutant is unresponsive to circadian phase changes induced by temperature and is arrhythmic if entrained to temperature steps (Salome and McClung 2005b). In contrast, it appears that TOC1 has a minor role in this mechanism as a toc1 mutant retains a wild-type entrainment response to temperature steps (Somers et al. 1998b). Further characterization of this sensitivity is dependent upon identification of temperature sensors in Arabidopsis.
Spatial Diversity in Clock Gene Function
Animal circadian clocks have long been recognized to contain a master clock which synchronizes multiple “slave” clocks in other tissues (reviewed by Aton and Herzog 2005). This hierarchical arrangement of the clock permits individual tissues to utilize subsets of circadian genes (DeBruyne et al. 2007a, b). In comparison, plants are thought to measure time using cell-autonomous circadian oscillators (Thain et al. 2000, 2002; Salome and McClung 2004), although it has remained unclear until recently whether each of these independent plant clocks share common core components across different cell types. It now appears that certain loops of the plant clock act predominantly in certain tissues. PRR3 has been shown to be predominantly expressed in Arabidopsis vasculature (Para et al. 2007) while recent microarray analysis has indicated that only a subset of genes known to have a circadian expression pattern in aerial tissues oscillate in hydroponically grown roots (James et al. 2008). Such data suggest that circadian rhythmicity in roots is controlled by a simplified mechanism and is dramatic evidence that plant circadian rhythms need not be controlled by a uniform set of components. In support of this concept, experiments using RNAi to reduce PRR3 mRNA levels induce a greatly pronounced shortening of the circadian clock when measured using vasculature-specific luciferase reporter constructs in comparison to those with a broader range of expression (Para et al. 2007). The use of modified clock circuitry in different plant tissues likely allows altered sensitivity to environmental stimuli and stresses and it will be interesting in the future to determine the functional role of tissue-specific circadian oscillations.
Future Directions
Our understanding of the Arabidopsis circadian clock at a transcriptional level has progressed rapidly, aided by the high-throughput capabilities of luciferase reporter mutant screens and microarray assays. While it is clear that a large percentage of the transcriptome is regulated by the circadian clock (Covington et al. 2008; Hazen et al. 2009), our understanding of the molecular processes underlying these large-scale changes in transcription remains limited. Recent work has identified a correlation between histone acetylation and transcriptional activity at the TOC1 locus (Perales and Mas 2007) and similarly, a degree of histone methylation is associated with changes in transcriptional activity at CCA1 and LHY loci (Ni et al. 2009). Both of these observations suggest that epigenetic marks may regulate circadian gene expression and identification of proteins responsible for these epigenetic marks will allow a more thorough understanding of transcriptional regulation by the clock.
While transcriptional regulation is clearly important for Arabidopsis clock function, it is also increasingly clear that the rhythms generated by the transcriptional clock are modulated by a range of posttranslational modifications. TOC1 and PRR proteins are differentially phosphorylated and degraded over the course of a day (Murakami-Kojima et al. 2002; Mas et al. 2003; Fujiwara et al. 2008) while ZTL protein accumulates with a circadian rhythm despite being transcribed at a regular rate (Somers et al. 2000; Kim et al. 2003). It is equally apparent that endogenous rhythms are regulated by changes in cytosolic composition, such as the concentration of free Ca2+ (Dodd et al. 2005a, 2007; Hastings et al. 2008). Considering these examples, it is unlikely that we have either identified all components of the Arabidopsis clock or that we yet fully understand the subtleties of action and regulation of characterized transcripts and proteins.
Ultimately, it will be important to transfer our understanding of the clock to real-world applications. Given the suggested role of the circadian clock in the regulation of plant responses to abiotic stresses (Walley et al. 2007; Mizuno and Yamashino 2008), it is possible that altering expression of certain clock components may confer enhanced stress tolerance. Indeed, Arabidopsis prr5 prr7 prr9 triple mutants have recently been reported to have an enhanced cold, drought, and salt tolerance, caused by increased expression of stress-responsive genes (Nakamichi et al. 2009). Such data are in agreement with work demonstrating that plants are differentially responsive to temperature over the course of a day and that this gating is controlled by the circadian clock (Fowler et al. 2005). Further work to understand the process by which stress response pathways and the circadian clock interact will likely be a fruitful course of investigation.
References
Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293(5531):880–883
Aton SJ, Herzog ED (2005) Come together, right & now: synchronization of rhythms in a mammalian circadian clock. Neuron 48(4):531–534
Briggs WR (2007) The LOV domain: a chromophore module servicing multiple photoreceptors. J Biomed Sci 14(4):499–504
Brunner M, Schafmeier T (2006) Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes Dev 20(9):1061–1074
Carre IA (2002) ELF3: a circadian safeguard to buffer effects of light. Trends Plant Sci 7(1):4–6
Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114(5):537–543
Christie JM (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58:21–45
Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5(8):e222
Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13(6):1305–1315
Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9(8):R130
Dardente H, Cermakian N (2007) Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol Int 24(2):195–213
DeBruyne JP, Weaver DR, Reppert SM (2007a) CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci 10(5):543–545
DeBruyne JP, Weaver DR, Reppert SM (2007b) Peripheral circadian oscillators require CLOCK. Curr Biol 17(14):R538–R539
Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12(12):2499–2510
Devlin PK, Kay SA (2001) Circadian photoperception. Annu Rev Physiol 63:677–694
Dodd AN, Love J, Webb AA (2005a) The plant clock shows its metal: circadian regulation of cytosolic free Ca(2+). Trends Plant Sci 10(1):15–21
Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA (2005b) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309(5734):630–633
Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie JM, Robertson FC, Jakobsen MK, Goncalves J, Sanders D, Webb AA (2007) The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318(5857):1789–1792
Dong G, Golden SS (2008) How a cyanobacterium tells time. Curr Opin Microbiol 11(6):541–546
Dowson-Day MJ, Millar AJ (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17(1):63–71
Dubruille R, Emery P (2008) A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol Neurobiol 38(2):129–145
Dunlap JC, Loros JJ, Liu Y, Crosthwaite SK (1999) Eukaryotic circadian systems: cycles in common. Genes Cells 4(1):1–10
Dunlap JC, Loros JJ, Colot HV, Mehra A, Belden WJ, Shi M, Hong CI, Larrondo LF, Baker CL, Chen CH, Schwerdtfeger C, Collopy PD, Gamsby JJ, Lambreghts R (2007) A circadian clock in Neurospora: how genes and proteins cooperate to produce a sustained, entrainable, and compensated biological oscillator with a period of about a day. Cold Spring Harb Symp Quant Biol 72:57–68
Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ (2005) Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170(1):387–400
Eriksson ME, Hanano S, Southern MM, Hall A, Millar AJ (2003) Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218(1):159–162
Fankhauser C, Staiger D (2002) Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216(1):1–16
Farre EM, Kay SA (2007) PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J 52(3):548–560
Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15(1):47–54
Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137(3):961–968
Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283(34):23073–23083
Green RM, Tingay S, Wang ZY, Tobin EM (2002) Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129(2):576–584
Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci U S A 99(17):11513–11518
Hall A, McWatters HG (2006) Endogenous plant rhythms, ed, vol. Blackwell, Oxford
Hardin PE (2005) The circadian timekeeping system of Drosophila. Curr Biol 15(17):R714–R722
Harmer S (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113
Harmer SL (2009) The circadian system in higher plants. Annu Rev Plant Biol 60(1):357–377
Harmon F, Imaizumi T, Gray WM (2008) CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J 55(4):568–579
Hastings MH, Maywood ES, O'Neill JS (2008) Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18(17):R805–R815
Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, Borevitz JO, Kay SA (2009) Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol 10(2):R17
Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11(11):550–558
James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG (2008) The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322(5909):1832–1835
Kiba T, Henriques R, Sakakibara H, Chua N-H (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19(8):2516–2530
Kim WY, Geng R, Somers DE (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci U S A 100(8):4933–4938
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449(7160):356–360
Li Q-H, Yang H-Q (2007) Cryptochrome signaling in plants. Photochem Photobiol 83(1):94–101
Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138(4):2374–2385
Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1(2005):0013
Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2:59
Martin-Tryon EL, Harmer SL (2008) XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell 20:1244–1259
Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143(1):473–486
Martinez-Garcia JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288(5467):859–863
Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426(6966):567–570
Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41(9):1002–1012
McClung CR (2006) Plant circadian rhythms. Plant Cell 18(4):792–803
McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408(6813):716–720
McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144:391–401
Michael TP, Salome PA, McClung CR (2003a) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci U S A 100(11):6878–6883
Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR (2003b) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302(5647):1049–1053
Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, Givan SA, Yanovsky M, Hong F, Kay SA, Chory J (2008a) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4(2):e14
Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J (2008b) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6(9):e225
Millar AJ (2004) Input signals to the plant circadian clock. J Exp Bot 55(395):277–283
Millar AJ, Short SR, Chua NH, Kay SA (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4(9):1075–1087
Mizuno T, Yamashino T (2008) Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol 49(3):481–487
Murakami-Kojima M, Nakamichi N, Yamashino T, Mizuno T (2002) The APRR3 component of the clock-associated APRR1/TOC1 quintet is phosphorylated by a novel protein kinase belonging to the WNK family, the gene for which is also transcribed rhythmically in Arabidopsis thaliana. Plant Cell Physiol 43(6):675–683
Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T (2005a) The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol 46(4):609–619
Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005b) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46(5):686–698
Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T (2007) Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol 48(6):822–832
Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50:447–462
Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457(7227):327–331
Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448(7151):358–361
Panda S, Hogenesch JB, Kay SA (2003) Circadian light input in plants, flies and mammals. Novartis Found Symp 253:73–82 discussion 82–78, 102–109, 281–104
Para A, Farre EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19(11):3462–3473
Perales M, Mas P (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19:2111–2123
Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323(5920):1481–1485
Rand DA, Shulgin BV, Salazar D, Millar AJ (2004) Design principles underlying circadian clocks. J R Soc Interface 1(1):119–130
Robertson FC, Skeffington AW, Gardner MJ, Webb AA (2009) Interactions between circadian and hormonal signalling in plants. Plant Mol Biol 69(4):419–427
Rockwell NC, Su Y-S, Lagarias JC (2006) Phytochrome structure and signalling mechanisms. Annu Rev Plant Biol 57(1):837–858
Salome PA, McClung CR (2004) The Arabidopsis thaliana clock. J Biol Rhythms 19(5):425–435
Salome PA, McClung CR (2005a) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17(3):791–803
Salome PA, McClung CR (2005b) What makes the Arabidopsis clock tick on time? A review on entrainment. Plant Cell Environ 28(1):21–38
Salome PA, Xie Q, McClung CR (2008) Circadian timekeeping during early Arabidopsis development. Plant Physiol 147(3):1110–1125
Sato E, Nakamichi N, Yamashino T, Mizuno T (2002) Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol 43(11):1374–1385
Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318(5848):261–265
Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13(12):2659–2670
Somers DE, Devlin PF, Kay SA (1998a) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282(5393):1488–1490
Somers DE, Webb AA, Pearson M, Kay SA (1998b) The short-period mutant, toc1–1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125(3):485–494
Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101(3):319–329
Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16(3):769–782
Somers DE, Fujiwara S, Kim WY, Suh SS (2007) Posttranslational photomodulation of circadian amplitude. Cold Spring Harb Symp Quant Biol 72:193–200
Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C (2003) The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev 17(2):256–268
Stelling J, Gilles ED, Doyle FJ 3rd (2004) Robustness properties of circadian clock architectures. Proc Natl Acad Sci U S A 101(36):13210–13215
Thain SC, Hall A, Millar AJ (2000) Functional independence of circadian clocks that regulate plant gene expression. Curr Biol 10(16):951–956
Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ (2002) The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol 130(1):102–110
Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet 3(10):1800–1812
Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93(7):1207–1217
Yakir E, Hilman D, Hassidim M, Green RM (2007) CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol 145(3):925–932
Zeilinger MN, Farre EM, Taylor SR, Kay SA, Doyle FJ 3rd (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2:58
Zhong HH, Painter JE, Salome PA, Straume M, McClung CR (1998) Imbibition, but not release from stratification, sets the circadian clock in Arabidopsis seedlings. Plant Cell 10(12):2005–2017
Acknowledgments
I thank Stacey Harmer, Cory Ellison, and Lauren Headland for critical reading of the manuscript and James Richardson for assistance with illustrations.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Jones, M.A. Entrainment of the Arabidopsis Circadian Clock. J. Plant Biol. 52, 202–209 (2009). https://doi.org/10.1007/s12374-009-9030-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-009-9030-1