Abstract
A rational four-step strategy to identify novel bacterial glycosyl hydrolases (GH), in combination with various fungal enzymes, was applied in order to develop tailored enzyme cocktails to efficiently hydrolyze pretreated lignocellulosic biomass. The fungal cellulases include cellobiohydrolase I (CBH I; GH family 7A), cellobiohydrolase II (CBH II; GH family 6A), endoglucanase I (EG I; GH family 7B), and β-glucosidase (βG; GH family 3). Bacterial endocellulases (LC1 and LC2; GH family 5), β-glucosidase (LβG; GH family 1), endoxylanases (LX1 and LX2; GH family 10), and β-xylosidase (LβX; GH family 52) from multiple sources were cloned, expressed, and purified. Enzymatic hydrolysis for varying enzyme combinations was carried out on ammonia fiber expansion (AFEX)-treated corn stover at three total protein loadings (i.e., 33, 16.5, and 11 mg enzyme/g glucan). The optimal mass ratio of enzymes necessary to maximize both glucan and xylan yields was determined using a suitable design of experiments. The optimal hybrid enzyme mixtures contained fungal cellulases (78% of total protein loading), which included CBH I (loading ranging between 9-51% of total enzyme), CBH II (9-51%), EG I (10-50%), and bacterial hemicellulases (22% of total protein loading) comprising of LX1 (13%) and LβX (9%). The hybrid mixture was effective at 50°C, pH 4.5 to maximize saccharification of AFEX-treated corn stover resulting in 95% glucan and 65% xylan conversion. This strategy of screening novel enzyme mixtures on pretreated lignocellulose would ultimately lead to the development of tailored enzyme cocktails that can hydrolyze plant cell walls efficiently and economically to produce cellulosic ethanol.
Similar content being viewed by others
Introduction
Using renewable resources for production of fuels and chemicals has attracted significant attention in recent years [1–3]. Lignocellulosic biomass provides a unique, low-cost, plentiful, and renewable resource for the sustainable production of biofuels [4, 5]. The utilization of cellulosic biomass as an industrial feedstock would result in the development of rural economies and diversify any nation’s energy portfolio [6–8]. It has been estimated that the amount of carbon fixed by plants is over 100 billion tons per year [9]. Among this fixed carbon, there is approximately 252 million tons of corn stover residues available in the USA each year, making it one of the most abundant agricultural feedstocks that can be used to produce cellulosic ethanol [10].
Lignocellulosics are comprised of a complex intermeshed matrix of cellulose, hemicellulose, and lignin [11, 12]. Successful conversion of lignocellulosic biomass to ethanol requires an efficient and economical pretreatment method, high sugar yields during enzymatic hydrolysis, and effective microbial fermentation of the hydrolyzed pentose and hexose sugars. Typically, the recalcitrant lignocellulosic matrix has to be thermochemically pretreated to increase the accessibility of cellulose and hemicellulose for subsequent enzymatic hydrolysis [2, 13]. Currently, the high costs of pretreatment and enzyme production are the major factors affecting the economics of lignocellulosic biorefineries [14].
In nature, both fungi and bacteria have their own unique machinery to deconstruct plant cell walls [15]. For certain fungi, a battery of cellulases, hemicellulases, and other accessory enzymes are extracellularly secreted to synergistically hydrolyze cell walls, while releasing monomeric and oligomeric sugars for fungal metabolism [16]. On the other hand, anaerobic bacterial enzymes are typically aggregated and assembled on a complex scaffold structure through various integrating modules known as cohesins and dockerins [17]. These enzyme complexes, known as cellulosomes, are attached to the surface of the bacterial cell walls [18–20]. Few studies have investigated the synergism among catalytic domains of various bacterial enzymes, and the synergistic interactions between bacterial and fungal hydrolases acting on pretreated lignocellulosic biomass. Some reports have shown exo/exo and exo/endo synergism between fungal and bacterial enzymes hydrolyzing crystalline cellulose [21, 22]. Recent publications have reported synergy between Trichoderma and Serratia/Streptomyces based on chitin-degrading hydrolases completely hydrolyzing untreated crab shells [23]. But, very few reports are available on the nature of synergistic interactions between bacterial and fungal enzymes, especially bacterial hemicellulases hydrolyzing pretreated lignocellulosic biomass.
In this study, we enzymatic digestibility of Ammonia Fiber Expansion (AFEX)-treated corn stover was evaluated by varying combinations of fungal and bacterial glycosyl hydrolases. Fungal enzymes (cellobiohydrolase (CBH) I, CBH II, and endoglucanase (EG I)) were purified from suitable commercial sources (Spezyme CP); while β-glucosidase (βG) was purified from Novozyme 188. Two cellulases (LC1 and LC2), two xylanases (LX1 and LX2), one β-glucosidase (LβG), and one β-xylosidase (LβX) were obtained from various bacterial sources (e.g., Clostridium, Geobacillus, and Dictyoglomus). This paper presents a rational four-step strategy for designing an optimal enzyme cocktail, based on enzymes from multiple sources, to efficiently hydrolyze pretreated lignocellulosic biomass to help ultimately decrease the cost of cellulosic ethanol.
Methods
AFEX Pretreatment
AFEX pretreatment of corn stover was carried out as described in our previous work [24]. Milled corn stover (particle size <100 μm) with 60% moisture (kilogram water/kilogram dry biomass) was transferred to a high-pressure Parr reactor. Liquid ammonia (1 kg of ammonia/kg of dry biomass) was slowly charged to the vessel. The reactor was maintained at 90°C for 5 min and then the pressure was released explosively (total residence time ∼30 min). The instantaneous pressure drop in the vessel caused the ammonia to vaporize, with an explosive decompression and resulted in considerable biomass fiber disruption. The pretreated material was left under the fume hood overnight to remove residual ammonia and then kept at 4°C until further use. The composition of milled AFEX corn stover, based on the National Renewable Energy Laboratory laboratory analytical procedures (NREL LAP) protocol [25], was found to be 34.4% glucan, 22.4% xylan, 4.2% arabinan, 0.6% mannan, 1.4% galactan, 3.8% uronyl, 11% lignin, and 5.6% acetyl content.
Discovery and Cloning of LX1, LX2, LβX, and LβG
Samples from Yellowstone National Park hot springs were obtained during sampling trips in September 2003 and September 2004. Enrichment cultures were grown in YTP-2 medium containing (per liter) 2.0 g yeast extract, 2.0 g tryptone, 2.0 g sodium pyruvate, 1.0 g KCl, 2.0 g KNO3, 2.0 g Na2HPO4.7H2O, 0.1 g MgSO4, 0.03 g CaCl2, and 2.0 ml clarified tomato juice. Enrichments were performed at 70°C in flasks agitated at 200 rpm. A number of aerobic cultures were purified by selection of individual colonies on plates containing the above medium and 16.0 g/liter agar. For preparation of genomic DNA, liter cultures were grown from a single colony in YTP-2 medium and collected by centrifugation. The cell concentrate was lysed using a combination of SDS and proteinase K, and genomic DNA was isolated using a phenol/chloroform extraction [26]. The genomic DNA was precipitated, treated with RNase to remove residual contaminating RNA, and fragmented by hydrodynamic shearing (HydroShear apparatus, Genomic Solutions, Ann Arbor, MI, USA) to generate fragments of either 3-5 kb or 10 kb. The fragments were purified on an agarose gel, end repaired, and ligated into a high-stability, low copy vector (pSMART-LCKan, Lucigen, Middleton, WI, USA). The recombinant plasmids were then used to transform E. cloni 10G ELITE electrocompetent (Lucigen) cells and screened on plates containing 4-methylumbelliferyl-β-d-xylopyranoside or 4-methylumbelliferyl-β-d-cellobioside (MUC). DNA inserts of the positive clones were sequenced, and the enzymes of interest were subcloned into pET28a and the resulting vectors used to transform BL21 (DE3) chemically competent cells. Amino acid sequences were deduced from the DNA sequences of the clones using ExPaSy translate tool (http://expasy.org/tools/dna.html), and confirmed by N-terminal sequencing/sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE).
Discovery and Cloning of LC1 and LC2
Dictyoglomus turgidum strain 6724T (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; German Collection of Microorganisms and Cell Cultures) bacterial cell concentrate was a kind gift of Dr. Frank T. Robb, Center of Marine Biotechnology, University of Maryland Biotechnology Institute. Clostridium thermocellum bacterial cell concentrate was a kind gift of Dr. Paul Weimer, United States Department of Agriculture, Agricultural Research Service, United States Dairy Forage Research Center, Madison, WI, USA. The cell concentrate from each strain was lysed using a combination of SDS and proteinase K, and genomic DNA was purified using a phenol/chloroform extraction methodology [26]. Genomic DNA was prepared as described in the previous section. The recombinant plasmids were then used to transform electrocompetent cells and screened on plates containing MUC. LC1 was obtained from screening of the D. turgidum library, and LC2 from screening of the C. thermocellum library. DNA inserts of the positive clones were sequenced, the enzymes of interest were subcloned into pET28a and the resulting vectors were used to transform BL21 (DE3) chemically competent cells [27]. Amino acid sequences were deduced from the DNA sequences of the clones using ExPaSy translate tool (http://expasy.org/tools/dna.html), and confirmed by N-terminal sequencing/SDS PAGE.
Enzyme Expression and Purification
Plasmids containing the specific protein genes (for LC 1, LC 2, LX1, LX 2, LβG, and LβX) with a 6× N-terminal His tag were transformed into Escherichia coli BL21 (DE3) cells for protein expression. A starter culture, inoculated from frozen stocks, was grown overnight by incubating at 37°C in kanamycin (30 µg/ml) + 0.4% glycerol. Flasks of LB soy + kanamycin + 0.4% glycerol were inoculated with the overnight culture and grown to an OD600 of 0.7-0.8, induced with 1 mM IPTG (isopropylthio-β-d-galactoside) and then incubated overnight (16-18 h) at 37°C. Cells were harvested by centrifugation (4,000 rpm, 30 min) and the supernatant removed. Cell pellets were resuspended in lysis buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl) and sonicated using sonics vibra cell large tip. The mixture was centrifuged at 12,000 rpm for 30 min to remove the cell debris. The supernatant was heated to 60°C for 20 min and centrifuged again at 12,000 rpm to further remove precipitated debris. The supernatant was filtered through 0.5 µm filter and applied on 40 ml bed volume HIS-select nickel affinity gel (Sigma, St. Louis, MO, USA) equilibrated in lysis buffer. The column was washed with ten column volumes of lysis buffer. The bound protein was eluted using six column volumes of elution buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 300 mM Imidazole) and verified on Pierce 4-20% SDS-PAGE (Thermo Fisher Scientific, Rockford, IL, USA). The Ni-pool was concentrated using Amicon Ultra-15 membrane, dialyzed against storage buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl and 20% Glycerol) and quantified using the Pierce Bradford assay kit (Thermo Fisher Scientific, Rockford, IL, USA) with bovine serum albumin as the standard.
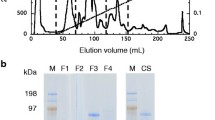
Fungal enzyme purification was performed using a fast protein liquid chromatography system (GE Healthcare, Buckinghamshire, United Kingdom) at room temperature, while the fraction collector was refrigerated. Cellulases (CBH I, CBH II, and EG I) and βG were purified from Spezyme CP (Genencor, Rochester, NY, USA) and Novo 188 (Novozyme, Davis, CA, USA), respectively. The purification methodology has been described in our previous work [28].
Strategy for Enzyme Screening on Realistic Lignocellulosic Substrates
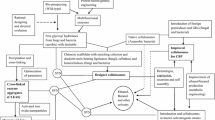
A simple four-step strategy was applied for screening and comparing activities of novel glycosyl hydrolases to develop enzyme mixtures that can efficiently saccharify pretreated lignocellulosic biomass (Fig. 1). A typical benchmark enzyme mixture could include fungal cellulases (CBH I + CBH II + EG I) along with a suitable βG. The goal was to compare the activity of novel enzymes with respect to a defined benchmark on realistic substrates like pretreated cellulosic biomass. The first step was to characterize the type of enzyme in order to assign it to a specific GH family (e.g., pNP-glycoside-based activity assays and glycosyl hydrolase family determination based on amino acid sequence similarity). The second step was doping the new enzyme/s along with the benchmark mixture to determine the effect on digestibility of pretreated biomass. It may be necessary to swap the corresponding type of enzyme from the benchmark mixture before adding new enzymes to compare relative improvements. This iterative method allows one to determine the most efficient enzyme/enzyme mixtures that have high activity on pretreated lignocellulosic biomass and avoid the pitfall of screening individual enzymes on unrealistic substrates (e.g., CMC, pNP-glycosides) [29]. Once a minimal enzyme mixture has been defined, it should be possible to further reduce enzyme dosage by optimizing the relative ratios of the enzymes in the mixture to maximize glucan and xylan digestibility [28].
Enzyme Activity Assays
The enzyme activity assays were based on a high-throughput microplate method as described in previous work [22]. A 2.2 ml deep-well microplate (Greiner, Monroe, NC, USA) was used to add 250 μl of 1% (w/v) stock substrate (CMC, Avicel, oat spelt xylan, cellobiose, xylobiose), 50 μl of 1 M citrate buffer (pH 5.0) and 200 μl of appropriately diluted enzyme samples (20 ng to 100 μg/well). The microplates were incubated at 50°C with shaking at 200 rpm for 10 min (cellobiose), 60 min (CMC, xylan), or 300 min (Avicel). The amount of glucose released was estimated using an enzyme-based glucose assay kit (R-Biopharm, Marshall, MI, USA). One unit of cellobiase activity was defined as one micromole of glucose released per milligram enzyme per minute under the assay conditions. For CMC-, Avicel-, and xylan-based substrates, the reducing sugars released were estimated using 3,5-dinitrosalicylic acid reagent [30]. One unit of CMCase, Avicelase, and xylanase activity was defined as 1 μmol of reducing sugars (as glucose equivalents) released per milligram enzyme per minute under the respective assay conditions.
The para-nitrophenyl (pNP)-based chromogenic substrates used were 4-nitrophenyl-β-d-cellobioside (pNPC), 4-nitrophenyl-β-d-glucopyranoside (pNPG), 4-nitrophenyl-β-d-xylopyranoside (pNPX), and 4-nitrophenyl-β-d-arabinofuranoside (pNPAf; Sigma-Aldrich, St. Louis, MO, USA). The assay mixtures containing 80 μl of 1 mM pNP substrate, 10 μl of 0.5 M citrate buffer (pH 5.0) and 10 μl of diluted enzymes (20 ng to 16 ug/well) in 350 μl micro plates were incubated at 50°C with shaking at 200 rpm. After 15 min reaction time, 200 μl of 1 M Na2CO3 was added to assay mixtures to arrest the hydrolytic reaction. The amount of pNP released was measured at OD420. One unit of enzyme activity was defined as 1 nanomol of p-nitrophenol released per milligram enzyme per minute under the assay conditions.
Enzymatic Hydrolysis of Pretreated Biomass
The hydrolysis experiments were performed in 2.2 ml deep well microplates (Greiner, Monroe, NC, USA) at 0.2% (w/w) total glucan loading in a total volume of 500 μl per well [24]. The enzyme and buffer mixtures were prepared separately and added simultaneously using a 96-channel automated pipette (JANUS, Perkin Elmer, Waltham). The hydrolysis experiments were performed using both Avicel and AFEX-treated corn stover as the substrates. The microplates were incubated at 50°C with shaking at 200 rpm for 24 h. The concentration of glucose and xylose in the hydrolyzates were measured using suitable enzyme-based assays. All experiments were carried out in duplicates.
Glucose and Xylose Assays
Glucose and xylose concentrations were measured using enzymatic kits purchased from R-Biopharm (Marshall, MI, USA) and Megazyme (Bray, Ireland), respectively. d-Glucose is first phosphorylated to d-glucose-6-phosphate using ATP and hexokinase. The reaction of d-glucose-6-phosphate with NADP+ is catalyzed by glucose-6-phosphate dehydrogenase to form d-gluconate-6-phosphate and NADPH. The reactions are stoichiometric to the amount of d-glucose and a corresponding increase in NADPH is measured at 340 nm to estimate the glucose concentration. The xylose assay is based on analogous two-step reactions. α-d-Xylose is converted to isomeric β-d-xylose by xylose mutarotase. β-d-Glucose-xylose is then reacted with NAD+ to form d-xylonic acid and NADH. The corresponding increase of NADH is measured at 340 nm to determine the xylose concentration. Suitable mixed glucose/xylose quality control (QC) standards were included during each assay to assess reliability of the method (less than 3-5% error deviation for known concentration of QC standards).
Results
Amino Acid Sequences of Bacterial Enzymes
The bacterial enzymes were isolated from thermophilic microbial enrichment isolates (See Methods Section and Table 1 for more details on the enzymes). BLAST analysis of all six sequences against UniprotKB database revealed 98-100% identity to known enzymes as listed in Table 1. The two cellulases (LC1 and LC2) are family 5 glycosyl hydrolases based on amino acid sequence similarity to D. turgidum- and C. thermocellum-based endoglucanases, respectively. The two hemicellulases (LX1 and LX2) are family 10 glycosyl hydrolases having sequence similarity to Geobacillus-based endoxylanases. The LβG and LβX belong to family 1- and 52-based glycosyl hydrolases with sequence similarity to Clostridium and Geobacillus-based enzymes, respectively.
Specific Activities of Bacterial and Fungal Enzymes
The enzymes were tested for their activity on different substrates at pH 5 (Table 2). LC1 and LC2 have significant pNP-cellobioside and CMC activity. Although both of the endocellulases were found to have significant activity on pNPC, their CMC activity was slightly lower or comparable to EG I. LX1 and LX2 were found to have much higher xylanase activity than EG I, though earlier work has reported another Geobacillus xylanase (with 88% similarity to LX1) to have lost 40-60% activity at pH 5 [31]. Recent work with a fungal endo-xylanase isolated from Trichoderma has shown that the Geobacillus enzyme has three- to fivefold higher activity on oat spelt xylan under identical conditions [28]. LβX was found to have high xylosidase activity but poor α-arabinofuranosidase activity, comparable to what has been reported earlier [32]. The bacterial LβG had significantly lower cellobiose activity than its fungal counterpart (βG). No noticeable Avicel activity was detected for any of the bacterial enzymes compared to the fungal cellulases.
Figure 2 depicts the hydrolysis yields on AFEX corn stover for all 6 bacterial enzymes added together as a mixture at varying pH and temperatures. The enzyme loading was 4 mg/g glucan each for LC1, LC2, LX1, and LX2; 2 mg/g glucan each for LβX and LβG. At pH 6.5 and 50°C, the xylose yield was approximately 50%, suggesting the high hemicellulase activity for the enzymes. However, the glucan conversion was significantly lower (<5%). The activity assays showed that none of the bacterial enzymes had any significant activity on Avicel (Table 2). Although the bacterial enzymes cloned belonged to thermophilic microbes, hydrolysis yields at 70°C were lower compared to 50°C. It is possible that the enzymes lost activity at high temperature during the prolonged incubation (24 h). Since the bacterial and fungal enzymes have a different working pH range, a mixture of both enzymes was tested on pH 6.5 and pH 4.5. For fungal enzymes, the optimal pH was found to be at 4.5-5 (data not shown). When tested under pH 6.5, significant loss in activity was observed for the fungal enzymes. For an equimass mixture of CBH I, CBH II, and EG I, the glucan hydrolysis yield decreased to 10% (pH 6.5) compared to 60% at pH 4.5 (24 h hydrolysis, data not shown).
Percent glucan (gray bar) and xylan (black bar) conversion after 24 h hydrolysis of AFEX-treated corn stover. Six bacterial enzymes were added together as a mixture at varying pH and temperature. Each enzyme mixture contains both bacterial cellulases and hemicellulases (4 mg/g glucan each for LC1, LC2, LX1, and LX2; 2 mg/g glucan each for LβX and LβG)
Interestingly, the bacterial β-xylosidase has been reported earlier to retain about 40% activity at pH 5 vs. pH 6.5 [32]. We noticed only a 25% loss in activity based on the overall xylan conversions for AFEX corn stover (Fig. 2). This would suggest that optimizing the bacterial hemicellulases at pH 4.5-5 along with fungal cellulases would be possible considering the significant retention in activity at acidic pH.
Doping and Swapping Bacterial/Fungal Cellulases
The experimental design for the doping and swapping experiments is shown in Table 3, which was conducted under specific assay conditions (pH 4.5-5.0, 50°C, 24 h) using AFEX-treated corn stover. From Fig. 3 I, experiments A-B show that swapping of LβG with βG does not significantly increase the glucose or xylose yield. From the results of experiment G, it can be observed that LβG showed lower glucan conversion even at much higher enzyme loadings (Fig. 3 II and Table 3). Experiments F-J also indicate that the bacterial β-glucosidase has much lower activity compared to its fungal counterpart. The current batch of bacterial enzymes does not possess substantial exo-cellulase activity to hydrolyze AFEX-treated corn stover. Therefore, purified fungal cellulases (CBH I, CBH II, and EG I) were doped into the enzyme mix in order to further enhance the glucose yield (experiments C-E). Doping any of the three fungal cellulases resulted in increasing the glucan conversion to around 20%. Interestingly, E has a higher xylose yield possibly due to cross-activity of EG I on xylan. Doping LC1, LC2, or both together into a fungal mixture did not significantly improve either the glucose or xylose hydrolysis yield (experiments K-M). The above results demonstrate that bacterial cellulases (LC1, LC2, and LβG) do not significantly improve the digestibility of pretreated biomass compared to fungal cellulases (CBH I, CBH II, and βG), despite the fact that the bacterial enzymes were found to have significant activity on artificial substrates (like CMC and pNP-glycosides). Wilson et al. have shown synergism between certain bacterial endo-glucanases and fungal exo-glucannases on filter paper, but the overall digestions were still quite low [33]. It is possible that due to lack of suitable exo-cellulase activity the bacterial cellulases (LC1 and LC2) are currently unable to completely hydrolyze the substrate. Previous results for C. thermocellum β-glucosidase have also revealed that gene functions encoding for hydrolytic activity on MU-glucoside and/or cellobiose are associated closely on the chromosome [34].
Percent glucan (gray bar) and xylan (black bar) conversions after 24 h hydrolysis of AFEX-treated corn stover. Enzyme mixtures used for experimental data sets in panel I, A-E; panel II, F-M; and panel III, H, O-R) are based on protein compositions listed in Table 3
Doping of Bacterial Hemicellulases to Fungal Benchmark Mixture
Although bacterial cellulases (LC1, LC2, and LβG) did not work effectively on their own or work synergistically with fungal enzymes, the bacterial hemicellulases (LX1, LX2, and LβX) were found to have significant activity on pretreated corn stover (experiments O-R). As shown in Fig. 3 III (experiment design shown in Table 3), doping LX1 and LX2 to the fungal benchmark mixture helps increase the xylose and glucose yield. Hydrolyzing xylan enhances the accessibility of the cellulases to the residual cellulose microfibrils; thereby, resulting in higher glucan conversions in the presence of suitable hemicellulases. When LX1 and LX2 were doped together (experiment Q), glucan conversion increased to 70% while no noticeable increase was seen for xylose (experiments O-P). Addition of LβX (experiment R) helped increase the xylan conversion substantially (71% glucan and 76% xylan conversion). In order to enhance both glucan and xylan conversion, LβX is important to hydrolyze soluble xylan-based oligosaccharides which are potential inhibitors of endoxylanases and cellulases. There have been reports on increased conversions on both xylan and glucan by supplementation of hemicellulases [35]. In order to completely digest the xylan fraction, β-xylosidase is indispensible to hydrolyze xylo-oligosaccharides, especially xylobiose to monomeric xylose [36]. Previous results have also shown that addition of bacterial hemicellulases to fungal cellulases results in increasing both glucose and xylose yields [37].
Enzyme Mixture Optimization
Previous results (Fig. 3) have demonstrated the synergistic interactions between fungal cellulases (CBH I, CBH II, EG I, and βG) and bacterial hemicellulases (LX1, LX2, and LβG). However, individual enzyme ratios need to be optimized to allow further increase in glucan and xylan conversions. In order to do this, 73 different enzyme combinations were tested in duplicates (standard deviations were less than 5% for the replicates) and the average hydrolysis yields for both glucan and xylan were determined for three different protein loadings (Table 4). βG was loaded at a 10% (mass ratio) of the total remaining enzymes to ensure complete hydrolysis of cellobiose [38]. CBH I and CBH II were added at more than 20% (total protein excluding βG) in all mixtures to ensure sufficient cellulase activity. EG I added was at least 10% of the mixture. Bacterial LX1 and LX2 together were always more than 5% of total enzyme loading while bacterial LβX was greater than 1%. All of the above constraints were based on the fact that cellobiohydrolase, endoglucanase, β-glucosidase, endoxylanase, and β-xylosidase are indispensible for an efficient enzyme cocktail [28]. Deconstruction of crystalline cellulose is the major limiting step towards complete hydrolysis, hence requiring significant amount of cellulase loading. It is clear that increasing the bacterial hemicellulases loading beyond 10 mg/g of glucan does not significant increase the xylan conversion. Using the current cocktail of enzymes, the xylan conversion could not exceed 75% conversion even when glucan conversion was over 90%. It is possible that other hemicellulases (e.g., α-arabinofuranosidase, α-glucuronidase) are necessary to further increase the xylan conversion. Without suitable complementary hemicellulases, xylan conversion is a bottleneck and increasing the total enzyme loading alone would not result in 100% xylan conversion.
Glucan and xylan hydrolysis results for all mixtures are shown in Fig. 4 as a scatter plot. The relative ratio of the individual enzymes significantly affected the overall sugar yield. At 10 mg/g glucan loading, the highest glucan conversion is 63.1% while the lowest is 20.6%. The highest xylan conversion is 52.3% while the lowest is 20.1%, at the same protein loading. Similarly, major differences in overall conversions can be seen for other protein loadings as well. One of the best mixtures resulting in the highest glucan and xylan conversions contained 19% CBH I, 9% CBH II, 50% EG I, 7.8% LX1, 5.3% LX2, and 9.0% LβX.
Glucan (x-axis) versus xylan (y-axis) conversion after 24 h hydrolysis of AFEX-treated corn stover. Hydrolysis results from three different total enzyme loadings (inclusive of CBH I + CBH II + EG I + LX 1 + LX 2 + LβX, as listed in Table 5) are depicted by green triangles (30 mg/g glucan), red squares (15 mg/g glucan), and blue diamonds (10 mg/g glucan). An additional loading of 3, 1.5, and 1 mg/g glucan of βG was supplemented in each
Relationship Between Glucan and Xylan Conversions
Glucan and xylan conversion for various combinations of enzymes at three different total enzyme loadings are shown in Fig. 4. The three clusters for different enzyme loadings demonstrate that at higher enzyme loading, the glucan and xylan conversion is generally higher. Another interesting phenomenon observed is that at higher enzyme loadings, the shape of the data point cluster is narrower. While at lower enzyme loadings, the data points are more scattered. By applying linear regression on xylan conversions vs. corresponding glucan conversion for various enzyme mixtures, a linear relationship of the two variables is confirmed (Table 5). The P values are close to 0, indicating that the linear relationship has statistical significance. When the total enzyme loading was increased (from 10 to 30 mg/g glucan) the coefficient of determination (R 2) value increases as well. This validates our visual interpretation of the shape of the data cluster at low vs. high enzyme loadings.
CBH I and CBH II are both indispensible for efficient hydrolysis. For all three varying enzyme loadings, if the mixture does not contain CBH I, the glucan conversions are quite low. When both CBH I and CBH II are included, higher glucan conversions are possible (>90% glucan conversion at 30 mg/g glucan enzyme loading). Xylan conversion is not obviously affected by the presence of either CBH I or II. Xylan conversion tends to be slightly higher (5-10%) at higher enzyme loadings when both CBH’s are present. LX1 has slightly higher specific activity compared to LX2 on AFEX-treated corn stover.
Figure 5 is helpful in visually summarizing the optimal regions of enzyme ratios for maximizing both glucan and xylan digestibility. Different clusters of data (based on Table 4) were separated based on the overall ratio of cellulases (I), xylanases (II), and βX (III). The three enzyme loadings were plotted as insets a, b, and c representing 10, 15, and 30 mg/g glucan enzyme loading, respectively. At high enzyme loadings, the higher glucan and xylan yielding data points aggregate closely compared to the lower enzyme loading. This suggests that glucan and xylan yields are more sensitive to individual enzyme ratios at lower enzyme loading. At high cellulase loading (94%), the hemicellulase loading is much lower and both glucan and xylan yields are relatively lower. This further confirms our previous assumption that in order to maximize glucan yield, higher xylan hydrolysis yields are desirable. On the other hand, for higher hemicellulase loading (>37%), xylan conversion is slightly lower while glucan conversions drop significantly. Similar trends for βX at around 9% loading of total protein mass ratio are seen as well. To achieve high conversions of glucan and xylan, 78% cellulases (CBH I, CBH II, and EG I), 13% xylanase (LX1 and LX2), and 9% βX seems to be optimal. The optimal cellulase loading (total of 78%; total protein mass basis) for both CBH I and CBH II ranges 9-51%; while EG I ranges 10-50%.
Glucan (x-axis) versus xylan (y-axis) conversion after 24 h hydrolysis of AFEX-treated corn stover for varying relative ratios of cellulases (panel I), xylanases (panel II), and β-xylosidase (panel III) at three different total enzyme loadings (a, b, and c correspond to 10, 15, and 30 mg/g glucan enzyme loading, respectively)
Discussion
It is interesting to note that the cellobiase activity for the bacterial enzyme (LβG) is significantly lower than its fungal counterpart (lower activity even at pH 6.5, data not shown). Previous work has reported anaerobic bacterial enzyme complexes to be easily inhibited by cellobiose [39], suggesting that cellulosomal activity on cellobiose is relatively poorer (compared to their fungal counterparts). It is also possible that due to preferred metabolism of cellobiose and the hydrolyzed oligomers after phosphorylation by the bacterial cell, the activity of β-glucosidase is relatively poor compared to their fungal counterparts [40].
One of the limitations of hydrolyzing the substrate at pH 5 is the relatively lower activity of these bacterial enzymes (40-50% of optimum at pH 6.5) reported by Bravman et al. [32]. This would suggest that in the presence of a suitable bacterial exo/endo-cellulase complex (that have an optimum activity at pH 6.5), it would be possible to further lower the total enzyme loading (and maximize glucan/xylan conversion). There have also been several reports on the activity of GH family 43 β-xylosidase, with very few publications on their GH 52 counterparts [32]. This study is one of the first that reports the activity of GH 52 β-xylosidases on pretreated lignocellulosic biomass.
In summary, we have examined the activity of both fungal and bacterial based enzyme mixtures on a realistic lignocellulosic substrate (i.e., AFEX-pretreated corn stover). The results indicate that certain fungal cellulases and bacterial hemicellulases work synergistically together to maximize glucan and xylan digestibility. Bacterial xylanases (LX1, LX2, and LβX) can increase both the glucan and xylan hydrolysis yield when added along with fungal cellulases. Optimized ratios for individual enzymes are obtained by examining 73 unique enzyme mixture combinations. Close to 90% glucan and 70% xylan conversion is achieved for the optimal enzyme combinations. There is a high linear correlation between glucose and xylose hydrolysis yield observed. Especially at high enzyme loading, this relationship is more obvious. This could be explained based on the cell wall structural organization and the effect of pretreatment on it. Within the untreated cell wall ultra-structure, cellulose fibrils are embedded among thin sheaths of hemicellulose [12]. After pretreatment, the cell wall structure is modified chemically and ultrastructurally, to substantially enhance enzyme accessibility [13, 41]. Unlike dilute acid pretreatment, AFEX does not hydrolyze and extract any hemicellulose from the cell wall. Therefore, hemicellulases are crucial not only to maximize hemicellulose hydrolysis but also help enhance glucan digestion. The current sets of bacterial hemicellulases are not sufficient to completely hydrolyze AFEX-treated corn stover hemicellulose. In order to further increase xylan conversion greater than 70%, other hemicellulases such as α-arabinofuranosidase and α-glucuronidases would be necessary.
Abbreviations
- AFEX:
-
ammonia fiber expansion
- GH:
-
glycosyl hydrolases
- CBH I:
-
cellobiohydrolase I
- CBH II:
-
cellobiohydrolase II
- EG I:
-
endoglucanase I
- βG:
-
β-glucosidase
- LC1 and LC2:
-
bacterial endocellulases
- LβG:
-
bacterial β-glucosidase
- LX1 and LX2:
-
bacterial endoxylanases
- LβX:
-
bacterial β-xylosidase
- pNPC:
-
p-Nitrophenyl β-d-cellobioside
- pNPL:
-
p-Nitrophenyl β-d-lactopyranoside
- pNPG:
-
p-Nitrophenyl β-d-glucopyranoside
- pNPAf:
-
p-Nitrophenyl α-l-arabinofuranoside
- pNPX:
-
p-Nitrophenyl β-d-xylopyranoside
- CMC:
-
carboxymethyl cellulose
References
Lynd LR, Cushman JH, Nichols RJ, Wyman CE (1991) Fuel ethanol from cellulosic biomass. Science 251:1318–1323
Jørgensen H, Kristensen JB, Felby C (2007) Enzymatic conversion of lignocellulose into fermentable sugars: challenges and opportunities. Biofuels, Bioproducts, Biorefining 1:119–134
Moreira N (2005) Growing expectations: new technology could turn fuel into a bumper crop. Sci News 168:218–220
Greene N (2004) Growing energy. How biofuels can help end America's oil dependence. Growing energy: how biofuels can help end America's oil dependence.vii+ 86
Fulton L, Howes T (2004) Biofuels for transport: an international perspective. Paris, pp 1-210
Dale BE (1999) Biobased industrial products: bioprocess engineering when cost really counts. Biotechnol Prog 15:775–776
Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM (2006) Ethanol can contribute to energy and environmental goals. Science 311:506–508
Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA et al (2006) The path forward for biofuels and biomaterials. Science 311:484–489
Ryu DDY, Mandels M (1980) Cellulases-biosynthesis and applications. Enzyme Microb Technol 2:91–102
Rooney T (1998) Lignocellulosic feedstock resource assessment. pp 123
Dale BE (1987) Lignocellulose conversion and the future of fermentation biotechnology. Trends Biotech 5:287–291
Bidlack J, Malone M, Benson R (1992) Molecular structure and component integration of secondary cell walls in plants. Proc Okla Acad Sci 72:51–56
Jeoh T, Ishizawa CI, Davis MF, Himmel ME, Adney WS, Johnson DK (2007) Cellulase digestibility of pretreated biomass is limited by cellulose accessibility. Biotechnol Bioeng 98:112–122
Dale BE, Leong CK, Pham TK, Esquivel VM, Rios I, Latimer VM (1996) Hydrolysis of lignocellulosics at low enzyme levels: application of the AFEX process. Bioresour Technol 56:111–116
Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
Wood TM, McCrae SI, Bhat KM (1989) The mechanism of fungal cellulase action—synergism betwwen enzyme components of penicillium-pinophilum cellulase in solubilizing hydrogen bond ordered cellulose. Biochem J 260:37–43
Demain AL, Newcomb M, Wu JHD (2005) Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154
Doi RH (2008) Cellulases of mesophilic microorganisms—cellulosome and noncellulosome producers. pp 267-279
Boisset C, Chanzy H, Henrissat B, Lamed R, Shoham Y, Bayer EA (1999) Digestion of crystalline cellulose substrates by the Clostridium thermocellum cellulosome: structural and morphological aspects. Biochem J 340:829–835
Bayer EA, Lamed R, White BA, Flint HJ (2008) From cellulosomes to cellulosomics. Chem Rec 8:364–377
Baker JO, Ehrman CI, Adney WS, Thomas SR, Himmel ME (1998) Hydrolysis of cellulose using ternary mixtures of purified celluloses. pp 395-403
Baker JO, Adney WS, Thomas SR, Nieves RA, Chou Y-C, Vinzant TB et al (1995) Synergism between purified bacterial and fungal cellulases. Enzyme Degrade Ins Carb 618:113–141
Donzelli BGG, Ostroff G, Harman GE (2003) Enhanced enzymatic hydrolysis of langostino shell chitin with mixtures of enzymes from bacterial and fungal sources. Carbohydr Res 338:1823–1833
Chundawat SPS, Balan V, Dale BE (2008) High-throughput microplate technique for enzymatic hydrolysis of lignocellulosic biomass. Biotechnol Bioeng 99:1281–1294
NREL (2004) Chemical Analysis and Testing (CAT) standard procedures. Available at:http://www.nrel.gov/biomass/analytical_procedures.html
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning, a laboratory manual
(2005) Novagen, pET system manual, 11th edn
Gao D, Chundawat SPS, Krishnan C, Balan V, Dale BE (2009) Mixture optimization of six core glycosyl hydrolases for maximizing saccharification of ammonia fiber expansion (AFEX)-pretreated corn stover. Bioresour Technol. doi:10.1016/j.biortech.2009.10.056
Kabel MA, van der Maarel MJEC, Klip G, Voragen AGJ, Schols HA (2006) Standard assays do not predict the efficiency of commercial cellulase preparations towards plant materials. Biotechnol Bioeng 93:56–63
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Khasin A, Alchanati I, Shoham Y (1993) Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl Environ Microbiol 59:1725–1730
Bravman T, Zolotnitsky G, Belakhov V, Shoham G, Henrissat B, Baasov T et al (2003) Detailed kinetic analysis of a family 52 glycoside hydrolase: a beta-xylosidase from Geobacillus stearothermophilus. Biochemistry 42:10528–10536
Irwin DC, Spezio M, Walker LP, Wilson DB (1993) Activity studies of 8 purified cellulases—specificity, synergism, and binding domain effects. Biotechnol Bioeng 42:1002–1013
Kadam S, Demain AL, Millet J, Beguin P, Aubert JP (1988) Molecular cloning of a gene for a thermostable β-glucosidase from Clostridium thermocellum into Escherichia coli. Enzyme Microb Technol 10:9–13
Selig MJ, Knoshaug EP, Adney WS, Himmel ME, Decker SR (2008) Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresour Technol 99:4997–5005
Polizeli MLTM, Rizzatti ACS, Monti R, Terenzi HF, Jorge JA, Amorim DS (2005) Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol 67:577–591
Viikari L, Alapuranen M, Puranen T, Vehmaanpera J, Siika-Aho M (2007) Thermostable enzymes in lignocellulose hydrolysis. Biofuels 108:121–145
Baker JO, Ehrman CI, Adney WS, Thomas SR, Himmel ME (1998) Hydrolysis of cellulose using ternary mixtures of purified cellulases. Appl Biochem Biotechnol 70–72:395–403
Steenbakkers PJM, Harhangi HR, Bosscher MW, van der Hooft MMC, Keltjens JT, van der Drift C et al (2003) Beta-glucosidase in cellulosome of the anaerobic fungus Piromyces sp strain E2 is a family 3 glycoside hydrolase. Biochem J 370:963–970
Zhang YHP, Lynd LR (2005) Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc Natl Acad Sci U S A 102:7321–7325
Mosier NS, Wyman CE, Dale BE, Elander RT, Lee YY, Holtzapple MT et al (2005) Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96:673–686
Acknowledgement
This work was funded by DOE Great Lakes Bioenergy Research Center (www.greatlakesbioenergy.org) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through Cooperative Agreement DE-FC02-07ER64494 between The Board of Regents of the University of Wisconsin System and the US Department of Energy. We also appreciate financial support, in initial stages of the project, from Michigan State Research Foundation (SPG grant). Special thanks to Genencor Division of Danisco (USA) and Novozyme Inc., for their generous gift of enzymes.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gao, D., Chundawat, S.P.S., Liu, T. et al. Strategy for Identification of Novel Fungal and Bacterial Glycosyl Hydrolase Hybrid Mixtures that can Efficiently Saccharify Pretreated Lignocellulosic Biomass. Bioenerg. Res. 3, 67–81 (2010). https://doi.org/10.1007/s12155-009-9066-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-009-9066-6