Abstract
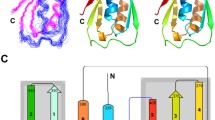
Pathogenic bacteria secrete pore-forming toxins (PFTs) to selectively defend against immune cells and to break through cellular barriers in the host. Understanding how PFTs attack cell membranes is not only essential for therapeutic intervention but for designing agents to deliver drugs to specific human cell subtypes, for example in anti-cancer or anti-viral therapies. Many toxins contain accessory domains that help recognize specific molecular epitopes on the membranes of target cells, including proteins, carbohydrates, and lipids. Here we report NMR assignments for the 94-residue 10 kDa C-terminal accessory domain of Bacillus cereus hemolysin II, HlyIIC, that has no known structural or functional homologues. The HlyIIC domain exists in a dynamic equilibrium due to cis/trans isomerization of its Gly86–Pro87 peptide bond. The cis and trans forms are about equally populated and are in slow exchange on the NMR timescale, giving rise to separate signals for approximately half of the residues in the domain. Assignments for the cis and trans forms were achieved with the aid of a P87M mutant that stabilizes the trans form, and separate sequential walks for the two forms in 3D NMR spectra of the wild-type HlyIIC. Based on backbone chemical shifts, the domain has a α1–α2–β1–β2–β3–β4–α3–β5 order of secondary structure elements. The last strand in the trans form and in the P87M mutant is shortened near Pro87 compared to the cis form. Both cis/trans isomerization and the P87M mutation cause large chemical shift changes throughout HlyIIC, suggesting that the proline is important in stabilizing the structure of the domain. The NMR assignments pave the way for solving the structures of the multiple conformational forms of HlyIIC and establishing their mechanism of interconversion.


Similar content being viewed by others
References
Case DA, Dyson HJ, Wright PE (1994) Use of chemical shifts and coupling constants in nuclear magnetic resonance structural studies on peptides and proteins. Methods Enzymol 239:392–416
Cheung MS, Maguire ML, Stevens TJ, Broadhurst RW (2010) DANGLE: a Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J Magn Reson 202(2):223–233
Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36(Web Server issue):W197–W201
Fisher CL, Pei GK (1997) Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23(4):570–571, 574
Marley J, Lu M, Bracken C (2001) A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR 20(1):71–75
Miles G, Bayley H, Cheley S (2002) Properties of Bacillus cereus hemolysin II: a heptameric transmembrane pore. Protein Sci 11(7):1813–1824
Provoda CJ, Lee KD (2000) Bacterial pore-forming hemolysins and their use in the cytosolic delivery of macromolecules. Adv Drug Deliv Rev 41(2):209–221
Shen Y, Bax A (2010) Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR 46(3):199–204
Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE (1996) Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274(5294):1859–1866
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59(4):687–696
Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD (1995) 1H, 13C and 15 N chemical shift referencing in biomolecular NMR. J Biomol NMR 6(2):135–140
Acknowledgments
This work was supported by a Project Grant from Wesleyan University to R.O. We thank Joyce Noble, Katie Kaus, and Naomi Hecht for their assistance with this project and Prof. Irina Russu for her generous guidance and advice.
Conflict of interest
The authors declare no conflict of interest.
Ethical standard
All experiments complied with all laws of the United States of America.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaplan, A.R., Maciejewski, M.W., Olson, R. et al. NMR assignments for the cis and trans forms of the hemolysin II C-terminal domain. Biomol NMR Assign 8, 419–423 (2014). https://doi.org/10.1007/s12104-013-9530-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-013-9530-2