Abstract
Cancer cells harbor unique mutations that theoretically create corresponding unique tumor-specific antigens. This class of mutated antigens represents an attractive target for cancer immunotherapy, but their identification has been cumbersome. By combining cancer genome sequencing with computational analysis of MHC binding, it is possible to predict and rank all of the possible mutated tumor antigens. This form of antigen screen is being combined with high throughput methods to measure the immune response to each candidate mutated antigen. Using these techniques, it is possible to systematically test each mutated tumor antigens for an associated immune response. Only a small fraction of the putative mutated antigens tested in this manner have been found to elicit an immune response, yet these responses appear to be both robust and durable. It is becoming increasingly clear that these mutated tumor antigens are an important target in the antitumor response. Studies incorporating this approach promise to improve our understanding of the inherent immunogenicity of individual cancers, potentially providing an explanation for the varying clinical responses to novel immunotherapeutic agents.
Similar content being viewed by others
The mapping of the cancer genome is revolutionizing our understanding of malignancy and its underlying complexity. The heterogeneity of cancer uncovered by comprehensive sequencing has highlighted the importance of personalized cancer therapy. At the same time, breakthroughs in tumor immunology are leading to the incorporation of cancer immunotherapy as a potent anti-cancer modality complementing traditional therapies. The results seen with anti-CTLA-4 and anti-PD-1/PD-L1 antibodies serve as a proof-of-concept that immunotherapy can achieve durable remissions with limited toxicity [1–5]. However, as with chemotherapy and targeted therapies, responses to immunotherapy are not universal. The deep and durable responses seen with immune checkpoint inhibitors are only elicited in a subset of patients and only in certain cancers [6]. In contrast to the advances in targeted therapy, we have as yet no effective a priori method of uniformly identifying potential immunotherapy responders. Furthermore, clinical and radiographic responses to immunotherapy can be delayed, making it difficult to identify responders even after treatment using standard criteria [7]. As comprehensive sequencing of individual cancers becomes more affordable and widely available, new methods are needed to translate genomic sequencing data into useful information to guide the burgeoning practice of immunotherapy. Coupled with an expanding armamentarium of immunomodulatory agents, further definition of the immunogenic landscape of cancer will be needed to unlock the full potential of cancer immunotherapy.
Native antigens versus neoantigens
Perhaps the greatest barrier to successful immunotherapy has been the lack of targetable tumor-associated antigens [8]. Because of their broad applicability, the most widely utilized strategies have been to target native tumor-associated antigens, such as cancer–testis antigens, which tend to be more commonly shared among different patients with the same malignancy [9–11]. While this allows therapy to be generalizable across patients, these native tumor-associated antigens are also found on germline/native tissues as well, resulting in immune tolerance that may be difficult to overcome and potentially off-target side effects. The response rates to native antigen-derived vaccines have been disappointing [10] with only a single vaccine receiving FDA approval [12].
The alternative approach is to instead target “neoantigens” formed by the somatic mutations unique to each patient’s tumor [13–15]. The unique mutational profile of each tumor is predicted to result in a corresponding unique set of neoantigens specific to that tumor. As compared to native antigens, neoantigens are less likely to experience central immunological tolerance, theoretically making them superior antigens. Neoantigens suffer from one major drawback; they are unique to each patient. Because their identification requires costly and extensive effort, individualized neoantigens have largely been avoided as immunotherapy targets. Yet, there is indirect evidence suggesting a central role of neoantigens in the endogenous antitumor response. A tumor-associated antigen screen in a long-term melanoma survivor found five of eight targeted tumor antigens were in fact neoantigens, with detectable responses lasting several years [16]. Furthermore, when tumor-infiltrating lymphocytes (TILs) from melanoma patients were stained with multimers for 145 melanoma epitopes, representing all known HLA-A2-restricted melanoma-associated native epitopes, <1 % of TILs were reactive [17]. These findings suggest that neoantigens may constitute the bulk of melanoma-associated antigens targeted by TILs.
Identification of neoantigens
The role of mutated tumor antigens as a target for the immune system had been theorized since the first experiments showing that syngeneic mice could reject transplanted carcinogen-induced tumors [18, 19]. The first identified tumor-specific antigens were idiotypes derived from the CDR regions of immunoglobulin variable domains found on lymphoma and myeloma cells [20]. These antigens are formed via natural immunoglobulin rearrangement machinery, creating unique epitopes with expression restricted to malignant cells. Their characterization was the first proof that mutated or rearranged genes could elicit antitumor responses which could be clinically exploited. However, the ability of idiotype vaccination to produce antitumor immune responses [21] did not translate into clinical success [22–24].
Whereas idiotype antigens created in the natural development of B cells were easily identified, neoantigens resulting from the underlying genetic instability of cancer proved more elusive. Decades after immune responses to idiotypes were first described, proof that mutated genes could produce targets for the antitumor immune response was demonstrated in murine models [25]. Later, autologous cytotoxic T lymphocytes isolated from human melanoma patients were found to target tumor-restricted mutated genes [26–28]. These studies, and those to follow, established neoantigens as a veritable class of tumor-associated antigens. However, these methods of neoantigen identification were laborious, largely relying on screens of TILs against cDNA tumor antigen libraries. Advancements in immunologic methods did little to facilitate the identification of these neoantigens, thus limiting their use in clinical applications.
In silico methods of neoantigen discovery
Next generation sequencing techniques are opening new avenues into neoantigen discovery. It is becoming increasingly affordable to sequence entire cancer genomes, allowing a personalized and complete characterization of the mutanome for each patient. Each non-synonymous mutation forms dozens of putative peptide targets. The number of potential neoantigens can be narrowed based on our understanding of antigen processing and presentation. To elicit a cytotoxic T cell response, neoantigens must be processed into peptides and presented on MHC class I molecules. Each HLA allele has a unique peptide-binding profile, with only a small fraction of potential peptides presented on any given HLA class I allele. Extremely accurate methods are now available to predict the binding affinity between any given peptide and specific HLA alleles [29, 30]. The netMHC model is frequently used and performs well in benchmarking studies, though it is simple to combine multiple models to improve performance [31, 32].
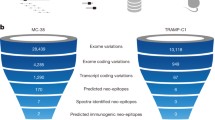
The mutation rate varies considerably from cancer to cancer and even within different cancer types [33–35]. The number of non-synonymous mutations range from about ten mutations per genome in acute myeloid leukemia to hundreds of mutations per genome in cancers such as melanoma and lung cancer [36–38]. Filtering this limited number of coding mutations through peptide-MHC affinity prediction algorithms would theoretically yield a manageable final number of potential neoantigens per tumor (Fig. 1).
This type of neoantigen prediction has been applied to the emerging cancer genome sequencing datasets, confirming a relatively small set of putative high-probability neoantigens. In the case of colon and breast cancer, there are an average of seven and 10 mutations per genome, respectively, which are predicted to form epitopes with high affinity binding to HLA-A0201 allele, one of the most common class I alleles [39]. Another study probed only missense mutations that had been annotated as functionally relevant in the catalogue of somatic mutations in cancer (COSMIC) database [40, 41]. A total of 26,672,189 corresponding peptides were tested against 57 human HLA-A and HLA-B alleles. Only 0.4 % of the peptides were predicted to have a high binding affinity to any human allele. As each patient has a total of only four HLA-A and HLA-B alleles, the frequency of peptides specific for self-MHC would be expected to be much lower. These studies suggest that mutations producing neoantigens with high MHC affinity are uncommon. While cancers with a high rate of mutation, such as melanoma, appear to have a significant number of relevant neoantigens, cancers with a lower mutation rate likely have few, if any, neoantigens.
The number of potential neoantigens depends on the threshold of peptide-MHC affinity used to establish candidacy. Higher affinity peptides induce stronger immune responses [42, 43], but the optimal cutoff is unclear. A recent model of murine tumor rejection found that peptide-MHC (pMHC) affinities of 10 nM or less are required for tumor rejection, whereas lower affinity peptides resulted in disease relapse [44]. Others have found that peptides with an IC50 as high as 200 nM were able to stimulate cytotoxic T cells to recognize and kill tumor cells in vitro [43]. In the case of neoantigen recognition in a melanoma patient, a neoantigen-specific response was seen with pMHC affinities up to 100 nM [45].
Using predicted peptide-MHC affinity in neoantigen screening requires knowledge of the corresponding individual’s HLA alleles. While conventional HLA serotyping or genotyping methods are available, these carry additional cost and labor and cannot be applied retrospectively to the large cancer genome datasets. In theory, the HLA type of the patient should be extractable from the genomic sequence. However, the high degree of polymorphism in the HLA locus poses significant challenges in identifying HLA alleles from short read sequences. New methods are available that demonstrate high accuracy in identifying HLA genotype from next generation sequencing data [46, 47]. The first large-scale analysis to incorporate HLA type extracted in this manner provides the most complete survey of the neoantigen landscape to date [48]. Using a more lenient cutoff pMHC affinity of 500 nM, the average number of neoantigens per genome for melanoma, renal cell carcinoma and chronic lymphocytic leukemia was found to be 488, 80 and 24, respectively.
Detecting the immune response to neoantigens
Combining cancer genome sequencing with peptide-MHC binding prediction analysis is useful in identifying candidate neoantigens, but how many of these actually produce an immune response? Using filtering of whole exome sequencing data, Robbins et al. [45] identified candidate neoantigens from melanoma patients who demonstrated near complete regression of disease after treatment with autologous TILs. The top 55 peptides with the highest predicted affinity to HLA-0201 were assessed, and it was found that four of these had elicited a measurable T-cell response. For each of the three patients analyzed, T cell responses as measured by IFN-gamma release could be detected against at least two neoantigens. The interferon-gamma responses against each neoantigen were almost as robust as the responses against the autologous tumor. Additionally, the proportion of reactive T cells was similar (~50 %) when stimulated with either neoantigen peptide or autologous tumor [45, 49]. T cells reactive against one such neoantigen were found to be persistent in the peripheral blood of one patient 5 years after transfer [49]. These findings suggest that a small fraction of predicted neoantigens may serve as the primary targets for melanoma-derived TILs. Thus, despite limiting the number of candidate neoantigens by peptide-MHC affinity, high throughput confirmatory methods will be needed to verify the small number of true neoantigen antitumor targets.
An alternative method to functional antitumor assays is to determine antigen specificity via pMHC multimer staining [50]. High throughput peptide-MHC production is possible through the use of a UV-cleavable loading peptide in the production of class I MHC tetramers [51] and combinatorial pMHC labeling which allows up to 64 distinct pMHCs to be assayed in the same sample [52, 53]. Peptide-MHC multimer staining represents an appealing methodology for rapid screening T cell responses against large numbers of candidate neoantigens. Van Rooij et al. [54] employed this strategy in a neoantigen screen of an ipilimumab-responsive melanoma patient. The high number of non-synonymous somatic mutations associated with this specific tumor allowed for further filtering of the whole exome data. In addition to predicting peptide-MHC affinity, the authors also used RNAseq data to account for gene expression and also filtered by predicted proteasome processing [55]. This yielded 448 candidate neoepitopes which were synthesized into pMHC multimers with combinatorial labeling. T cell responses were detected against two of these neoepitopes. One peptide was specific for 0.003 % of CD8-positive TILs, while the other stained for 3.3 % of CD8-positive TILs. By using multimers, the neoantigen-specific response could be easily tracked in serial samples. This approach enables an efficient and comprehensive cataloguing of all the putative neoantigens for a particular cancer. Similar to the findings of Robbins et al., their findings suggest that only a small number of candidate neoantigen actually elicit an immune response.
These methods of neoantigen screening do have inherent limitations. They assume presentation of a mutated neoantigen peptide from genomic sequencing data, but this assumption may not hold true in all cases. Allelic expression may occur preferentially or exclusively from a preserved germline allele instead of the mutated allele, resulting in null expression of the putative neoantigen. This possibility can be partially addressed through verification of mutant allele expression, either by using RNAseq as the initially sequencing method or else through allele-specific expression measurement by a method such as quantitative allele-specific polymerase chain reaction. Additionally, cancer cells may evade detection via alteration of their antigen processing machinery or through downregulation of MHC class I expression, rendering the neoantigen undetectable by the immune system. Verifying the expression of MHC class I on the surface of the tumor cells can help exclude this possibility. However, confirming a neoantigen-specific tumor response with certainty would require directly testing reactivity of sorted neoantigen-specific cells against autologous tumor cells.
The use peptide-MHC multimers to detect the immune response also poses challenges. One limitation is that multimers must be synthesized for any pMHC to be tested, and uncommon or rare HLA alleles may be difficult to study. This is alleviated to some degree by the observation that groups of MHC class I alleles, called supertypes, display similar affinities for peptides and that algorithms predicting peptide-MHC affinity can be applied across members within the same supertype [56]. Even alleles in different supertypes sometimes demonstrate shared affinities [57]. Similarly, T cell receptors can demonstrate promiscuity in recognizing peptides in the context of multiple MHC alleles as well [58]. Another limitation to the use of pMHC multimers is that their application to MHC class II analysis is substantially more troublesome. This is in part due to the inferiority of the algorithms for predicting affinity for peptides binding MHC class II [59], though many models do exist. Another difficulty is that peptide-specific CD4 cells have lower affinity for their pMHC target and are present at lower frequencies [60, 61], thus complicating their detection. Thus far, the role of MHC class II-restricted neoantigens has not been investigated, despite mounting evidence of the importance of the CD4 T cell population in the antitumor immune response [62].
Immunoediting and implications for neoantigens
There are theoretical considerations that may explain the rarity of immunogenic neoantigens found upon screening. The immune system undergoes constant surveillance of developing tumors, and thus, it is thought that immunogenic mutations are deleted during the development of tumors. This process has been dubbed immunoediting [63, 64], and now has been confirmed experimentally in two murine models. In the first, an oncogene-driven, endogenous tumor engineered to express neoantigens was found to undergo deletion of the neoantigens only when passaged in an immunocompetent background [65]. The second study used an “unedited” carcinogen-induced immunogenic sarcoma cell line generated in an immunodeficient background [66]. The tumor cells were sequenced and mutations were subjected to MHC class I peptide prediction algorithms to identify potential neoantigens. When injected into immunocompetent mice, the tumors were rejected in about 80 % of recipients. An immunodominant neoantigen was discovered in the unedited, parental tumors which elicited a T cell response in mice challenged with the parental tumor. When tumors that escaped rejection were resequenced, this mutation was deleted. Thus, passage through an immunocompetent host was shown to “edit” the immunogenic neoantigen, resulting in outgrowth of a subclone of tumor lacking the neoantigen. It would be expected that human tumors may edit immunogenic neoantigens in a similar manner, potentially explaining the low numbers such neoantigens seen in previous studies.
The effect of tumor immunoediting on neoantigen expression takes on increased importance when combined with our evolving understanding of intratumoral heterogeneity. Tumors are composed of a number of subclones with distinct genetic alterations [67–71]. Furthermore, comparison of primary tumors with sites of metastases show that each metastasis contains a mixture of mutations found in the primary lesion and a new set of mutations present only in the metastasis [69, 72], with the heterogeneity primarily due to the accumulation of passenger mutations [73]. This has important implications for the use of neoantigens as targets. Since neoantigen expression may be limited to a subclonal population, neoantigens associated with passenger mutations may be more easily edited, resulting in the outgrowth of a resistant clone lacking that mutation. Similarly, the allelic frequency of a given mutation may be an important consideration in the selection of target neoantigens, with high allelic frequency mutations representing superior targets.
An alternative strategy to unbiased neoantigen screening is to focus on neoantigens formed by founder or driver mutations. Natural immune responses are occasionally seen against these mutated proteins [26, 74, 75]. This strategy has been employed in vaccine development against frequently mutated genes such as ras, p53 and BCR-ABL with limited success [76–79]. Castle et al. [80] applied peptide-MHC affinity predictions to the B16F10 melanoma cell line with a focus on only those neoantigens associated with potential driver genes. Sequencing of the murine B16F10 melanoma cell line found 563 mutations, with several involving potential driver genes. Fifty mutated peptides were chosen that were related to potential driver genes and displayed high affinity for class I MHC. One-third of these peptides generated an immune response, and immunization with those peptides conferred a protective effect against tumor inoculation. A similar analysis was performed using the COSMIC database for human mutations [81]. Mutations which were present in 5 % or greater of cancers were subjected to HLA-binding prediction algorithms to find neoantigens with affinity to class I HLA alleles. Thirty-six mutations were tested against all HLA class I alleles and candidates were weighted by frequency of the mutation in a given cancer, binding affinity of the peptide for a given HLA allele and the frequency of that allele in the general population. Interestingly, the top six candidates by this methodology were all KRAS mutations. Since the HLA type was factored only as an allelic frequency in the general population, it is possible that none of the mutations discovered occurred in a patient with a matching HLA allele.
Given the small number of driver mutations per tumor genome, restricting analysis to this subset of neoantigens may not be feasible nor may it be necessary. Those neoantigens discovered by sequencing and peptide-MHC affinity prediction have already survived the immunoediting process. Their persistence implies that either they contribute to tumor survival or their creation preceded a driver mutation that contributes to survival, allowing them to resist deletion. Tumors with these mutations may escape immune regulation via a number of alternatives to clonal deletion, including downregulation of antigen expression, downregulation of MHC molecules, dysfunction in antigen processing machinery or immune suppression through a number of mechanisms of immune dysregulation [82]. These cells may be the ones most amenable to cancer immunotherapy, such as immune checkpoint blockade.
Clinical implications
The identification of specific neoantigens has a number of direct clinical implications. One obvious benefit would be in the design of tumor-specific vaccines. For the reasons discussed above, neoantigen-derived vaccines would theoretically be less prone to immune tolerance than currently used tumor vaccines, and immune responses to vaccines could be readily measured. Knowledge of specific neoantigens could also address several challenges facing the next generation of immunomodulatory agents in development, such as antibodies targeting PD1/PD-L1 and CD137. While these agents have produced some dramatic results in clinical trials, responses have been limited to a subset of patients with no clear biomarkers identified to aid in predicting response. Information regarding the presence of tumor-specific T cells and, perhaps more importantly, the immunophenotype of these tumor-specific T cells may help identify those patients most likely to benefit from immunomodulatory therapy. For example, it is known that PD-L1 expression by tumor cells does not accurately predict response to immune checkpoint blockade [4], indicating that perhaps the phenotype of the tumor-specific lymphocytes may be a better predictor than the tumor itself. One possibility is that PD-1 expression (a marker of lymphocyte exhaustion) on neoantigen-specific T cells may better predict response to anti-PD-1/PD-L1 antibody therapy. Additionally, monitoring the neoantigen-specific tumor response after immunomodulatory therapy may provide a superior assessment of response than conventional imaging methods, which are unable to differentiate progressive disease from early immune responses, a phenomenon termed pseudoprogression. Lastly, identification of neoantigen-specific lymphocytes could improve the production protocols used for adoptive immunotherapy from TILs. As an alternative approach to reversing T cell exhaustion with immune checkpoint inhibitors, T cell receptors (TCRs) from neoantigen-specific cells could easily be cloned and transduced into lymphocytes. There is evidence that transduction of tumor-specific TCRs into naïve cells may produce a more effective population of T cells for adoptive immunotherapy [83].
Conclusion
Cancer genome sequencing promises to open the once-hidden field of neoantigens to investigation. The challenge remains, how to translate the genomic and epigenetic landscapes of cancer into an understanding of the immunogenicity of cancer. Identification of neoantigens through sequencing and computational methods has the potential to transform our approach to immunotherapy. This information could be used to predict those patients most likely to respond to immunotherapy or to detect early responses to therapy prior to traditional clinical indicators. It may also be the key to the development of successful cancer vaccines, with trials already being planned [84]. The next years will reveal whether neoantigens will finally provide cancer immunotherapy with the elusive tumor antigens needed to propel the field.
References
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–65.
Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369(2):134–44.
Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54.
Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–33.
Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–23.
Ribas A. Tumor immunotherapy directed at PD-1. N Engl J Med. 2012;366(26):2517–9.
Wolchok JD, Hoos A, O’Day S, Weber JS, Hamid O, Lebbe C, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15(23):7412–20.
Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15(17):5323–37.
Parmiani G, Castelli C, Dalerba P, Mortarini R, Rivoltini L, Marincola FM, et al. Cancer immunotherapy with peptide-based vaccines: what have we achieved? Where are we going? J Natl Cancer Inst. 2002;94(11):805–18.
Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10(9):909–15.
Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–25.
Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363(5):411–22.
Heemskerk B, Kvistborg P, Schumacher TN. The cancer antigenome. EMBO J. 2013;32(2):194–203.
Sensi M, Anichini A. Unique tumor antigens: evidence for immune control of genome integrity and immunogenic targets for T cell-mediated patient-specific immunotherapy. Clin Cancer Res. 2006;12(17):5023–32.
Rammensee HG, Singh-Jasuja H. HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev Vaccines. 2013;12(10):1211–7.
Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA. 2005;102(44):16013–8.
Kvistborg P, Shu CJ, Heemskerk B, Fankhauser M, Thrue CA, Toebes M, et al. TIL therapy broadens the tumor-reactive CD8(+) T cell compartment in melanoma patients. Oncoimmunology. 2012;1(4):409–18.
Gross L. Intradermal immunization of C3H mice against a sarcoma that originated in an animal of the same line. Cancer Res. 1943;3(5):326–33.
Foley EJ. Antigenic properties of methylcholanthrene-induced tumors in mice of the strain of origin. Cancer Res. 1953;13(12):835–7.
Lynch RG, Graff RJ, Sirisinha S, Simms ES, Eisen HN. Myeloma proteins as tumor-specific transplantation antigens. Proc Natl Acad Sci USA. 1972;69(6):1540–4.
Kwak LW, Campbell MJ, Czerwinski DK, Hart S, Miller RA, Levy R. Induction of immune responses in patients with B-cell lymphoma against the surface-immunoglobulin idiotype expressed by their tumors. N Engl J Med. 1992;327(17):1209–15.
Freedman A, Neelapu SS, Nichols C, Robertson MJ, Djulbegovic B, Winter JN, et al. Placebo-controlled phase III trial of patient-specific immunotherapy with mitumprotimut-T and granulocyte-macrophage colony-stimulating factor after rituximab in patients with follicular lymphoma. J Clin Oncol. 2009;27(18):3036–43.
Schuster SJ, Neelapu SS, Gause BL, Janik JE, Muggia FM, Gockerman JP, et al. Vaccination with patient-specific tumor-derived antigen in first remission improves disease-free survival in follicular lymphoma. J Clin Oncol. 2011;29(20):2787–94.
Levy R, Robertson M, Ganjoo K, JP L, J V, D D, editors. Results of a Phase 3 trial evaluating safety and efficacy of specific immunotherapy, recombinant idiotype (Id) conjugated to KLH (Id-KLH) with GM-CSF, compared to non-specific immunotherapy, KLH with GM-CSF, in patients with follicular non-Hodgkin’s lymphoma (fNHL). Proceedings of the 99th Annual Meeting of the American Association for Cancer Research; 2008; San Diego, CA: AACR.
Lurquin C, Van Pel A, Mariamé B, De Plaen E, Szikora JP, Janssens C, et al. Structure of the gene of tum- transplantation antigen P91A: the mutated exon encodes a peptide recognized with Ld by cytolytic T cells. Cell. 1989;58(2):293–303.
Wölfel T, Hauer M, Schneider J, Serrano M, Wölfel C, Klehmann-Hieb E, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269(5228):1281–4.
Coulie PG, Lehmann F, Lethé B, Herman J, Lurquin C, Andrawiss M, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci USA. 1995;92(17):7976–80.
Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183(3):1185–92.
Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 2008;9:8.
Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40(Web Server issue):W525–30.
Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24(11):1397–8.
Trost B, Bickis M, Kusalik A. Strength in numbers: achieving greater accuracy in MHC-I binding prediction by combining the results from multiple prediction tools. Immunome Res. 2007;3:5.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9.
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8.
Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–34.
Network CGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74.
Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43(5):442–6.
Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889–92.
Khalili JS, Hanson RW, Szallasi Z. In silico prediction of tumor antigens derived from functional missense mutations of the cancer gene census. Oncoimmunology. 2012;1(8):1281–9.
Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50.
Parkhurst MR, Salgaller ML, Southwood S, Robbins PF, Sette A, Rosenberg SA, et al. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol. 1996;157(6):2539–48.
Keogh E, Fikes J, Southwood S, Celis E, Chesnut R, Sette A. Identification of new epitopes from four different tumor-associated antigens: recognition of naturally processed epitopes correlates with HLA-A*0201-binding affinity. J Immunol. 2001;167(2):787–96.
Engels B, Engelhard VH, Sidney J, Sette A, Binder DC, Liu RB, et al. Relapse or eradication of cancer is predicted by peptide-major histocompatibility complex affinity. Cancer Cell. 2013;23(4):516–26.
Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–52.
Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, Su LF, et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc Natl Acad Sci USA. 2012;109(22):8676–81.
Liu C, Yang X, Duffy B, Mohanakumar T, Mitra RD, Zody MC, et al. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 2013;41(14):e142.
Rajasagi M, Shukla S, Fritsch E, Deluca D, Getz G, Hacohen N, et al., editors. Tumor Neoantigens Are Abundant Across Cancers. New Orleans: American Society of Hematology; 2013.
Lu YC, Yao X, Li YF, El-Gamil M, Dudley ME, Yang JC, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190(12):6034–42.
Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–6.
Rodenko B, Toebes M, Hadrup SR, van Esch WJ, Molenaar AM, Schumacher TN, et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat Protoc. 2006;1(3):1120–32.
Hadrup SR, Bakker AH, Shu CJ, Andersen RS, van Veluw J, Hombrink P, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat Methods. 2009;6(7):520–6.
Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods. 2009;6(7):497–9.
van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31(32):e439–42.
Nielsen M, Lundegaard C, Lund O, Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57(1–2):33–41.
Sette A, Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol. 1998;10(4):478–82.
Rao X, Hoof I, Costa AI, van Baarle D, Kesmir C. HLA class I allele promiscuity revisited. Immunogenetics. 2011;63(11):691–701.
Fleischhauer K, Tanzarella S, Wallny HJ, Bordignon C, Traversari C. Multiple HLA-A alleles can present an immunodominant peptide of the human melanoma antigen Melan-A/MART-1 to a peptide-specific HLA-A*0201+ cytotoxic T cell line. J Immunol. 1996;157(2):787–97.
Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130(3):319–28.
Nepom GT. MHC class II tetramers. J Immunol. 2012;188(6):2477–82.
Vollers SS, Stern LJ. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology. 2008;123(3):305–13.
Kennedy R, Celis E. Multiple roles for CD4+ T cells in anti-tumor immune responses. Immunol Rev. 2008;222:129–44.
Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–8.
Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331(6024):1565–70.
DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482(7385):405–9.
Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400–4.
Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–13.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92.
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–95.
Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–7.
Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–13.
Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13.
Kubuschok B, Neumann F, Breit R, Sester M, Schormann C, Wagner C, et al. Naturally occurring T-cell response against mutated p21 ras oncoprotein in pancreatic cancer. Clin Cancer Res. 2006;12(4):1365–72.
Cai A, Keskin DB, DeLuca DS, Alonso A, Zhang W, Zhang GL, et al. Mutated BCR-ABL generates immunogenic T-cell epitopes in CML patients. Clin Cancer Res. 2012;18(20):5761–72.
Gjertsen MK, Bakka A, Breivik J, Saeterdal I, Solheim BG, Soreide O, et al. Vaccination with mutant ras peptides and induction of T-cell responsiveness in pancreatic carcinoma patients carrying the corresponding RAS mutation. Lancet. 1995;346(8987):1399–400.
Pinilla-Ibarz J, Cathcart K, Korontsvit T, Soignet S, Bocchia M, Caggiano J, et al. Vaccination of patients with chronic myelogenous leukemia with bcr-abl oncogene breakpoint fusion peptides generates specific immune responses. Blood. 2000;95(5):1781–7.
Carbone DP, Ciernik IF, Kelley MJ, Smith MC, Nadaf S, Kavanaugh D, et al. Immunization with mutant p53- and K-ras-derived peptides in cancer patients: immune response and clinical outcome. J Clin Oncol. 2005;23(22):5099–107.
Rojas JM, Knight K, Wang L, Clark RE. Clinical evaluation of BCR-ABL peptide immunisation in chronic myeloid leukaemia: results of the EPIC study. Leukemia. 2007;21(11):2287–95.
Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72(5):1081–91.
Warren RL, Holt RA. A census of predicted mutational epitopes suitable for immunologic cancer control. Hum Immunol. 2010;71(3):245–54.
Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6(10):715–27.
Hinrichs CS, Borman ZA, Cassard L, Gattinoni L, Spolski R, Yu Z, et al. Adoptively transferred effector cells derived from naive rather than central memory CD8+ T cells mediate superior antitumor immunity. Proc Natl Acad Sci USA. 2009;106(41):17469–74.
Ott P. A phase I study with a personalized NeoAntigen cancer vaccine in Melanoma 2013.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khodadoust, M.S., Alizadeh, A.A. Tumor antigen discovery through translation of the cancer genome. Immunol Res 58, 292–299 (2014). https://doi.org/10.1007/s12026-014-8505-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-014-8505-4