Abstract
This paper describes a pilot query interface that has been constructed to help us explore a “concept-based” approach for searching the Neuroscience Information Framework (NIF). The query interface is concept-based in the sense that the search terms submitted through the interface are selected from a standardized vocabulary of terms (concepts) that are structured in the form of an ontology. The NIF contains three primary resources: the NIF Resource Registry, the NIF Document Archive, and the NIF Database Mediator. These NIF resources are very different in their nature and therefore pose challenges when designing a single interface from which searches can be automatically launched against all three resources simultaneously. The paper first discusses briefly several background issues involving the use of standardized biomedical vocabularies in biomedical information retrieval, and then presents a detailed example that illustrates how the pilot concept-based query interface operates. The paper concludes by discussing certain lessons learned in the development of the current version of the interface.
Similar content being viewed by others
Introduction
This paper describes a pilot query interface that has been constructed for searching the Neuroscience Information Framework (NIF). The query interface is “concept-based” in the sense that the search terms submitted through the interface must be selected from a standardized vocabulary of terms (concepts) that are structured in the form of the NIF Standardized (NIFSTD) ontology (Bug et al. 2008) that defines each concept and specifies relationships among the concepts. As a result, this concept-based query interface (CBQI) differs from a search tools such as Google, since Google allows free text (i.e., arbitrary words or phrases) to be entered as search terms.
One advantage of using a concept-based approach is that it has the potential to help resolve the naming heterogeneity that occurs when the identical concept is described using different terms in different neuroscience resources. The approach may also facilitate integration of neuroscience knowledge with future informatics advances, for example involving the use of ontologies and the semantic web, in biomedicine as a whole.
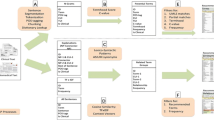
The construction of the NIF is an ongoing multi-institutional project (Gardner et al. 2008), supported by NIH as one of its Neuroscience Blueprint initiatives, whose goal is to help neuroscientists discover and access information available on the Web that is relevant to a neuroscience question of interest. The NIF contains three primary resources: the NIF Resource Registry, the NIF Document Archive, and the NIF Database Mediator (see Fig. 1).
-
The NIF Resource Registry is a database containing information about a wide range of different types of databases and other Web-based resources relevant to the neurosciences. For each resource, the Registry includes (1) a short text description of the resource, (2) contact information, (3) a URL pointer to the resource itself, and (4) a list of terms (that are mapped to NIFSTD concepts) that index/characterize the contents of the resource. This concept-based indexing is done at a high level of abstraction. Thus a resource containing data about neurons would be indexed using the concept “Neuron” with no further detail as to which specific types of neurons might be described within that resource. When performing a search of the NIF Registry, only these quite superficial descriptions can be searched by the CBQI. The contents of the resources themselves cannot be searched by the NIF Registry, but would need to be searched manually by the user after using the URL to link to the resource itself.
-
The NIF Document Archive is a repository of neuroscience articles and documents whose contents have been comprehensively indexed to facilitate rapid textual searching, using text words and phrases (Müller et al. 2008).
-
The NIF Database Mediator (Gupta et al. 2008) allows automated searching of the contents of a set of mediated databases whose internal vocabularies have been mapped to the NIFSTD ontology.
A schematic outline that shows the major components of the pilot NIF CBQI. The user enters a query into the query interface which passes that query to three components, each of which passes its search results back to the query interface, and then presents those results to the user, as described in detail “The Pilot NIF CBQI: an Example Search”
As described above, these three NIF resources are very different in their nature and in the type of search that each is designed to support. These differences pose challenges when designing a single interface from which searches can be automatically launched against all three resources simultaneously.
This paper first briefly outlines several background issues involving the use of standardized biomedical vocabularies/ontologies and their use in biomedical information retrieval. The paper then presents a detailed example that illustrates how the current pilot NIF CBQI operates. Finally the paper discusses lessons learned in the development of the current interface.
Background
There have been numerous efforts to develop standardized biomedical vocabularies designed to fulfill many different purposes. For example, one well-known vocabulary is the set of Medical Subheadings (MeSH) used by Medline to index the biomedical literature (http://www.nlm.nih.gov/mesh). Other vocabularies have focused on indexing clinical data, for example the Systematic Nomenclature of Medicine (SNOMED) (http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html). More recently a spectrum of vocabularies have been developed to index the biosciences, for example the Gene Ontology (Harris et al. 2004) and the Open Biomedical Ontologies (www.obofoundry.org).
A more broadly focused initiative is the Unified Medical Language System (UMLS) (http://www.nlm.nih.gov/research/umls/umlsmain.html) built and maintained by the National Library of Medicine (NLM). One goal of the UMLS is to provide a kind of unifying “Rosetta stone” for many of the diverse biomedical vocabularies that are in use for different purposes. The UMLS contains a metathesaurus of concepts (uniquely defined terms, e.g., “Neurons.” “Purkinje Cells”) to which terms used in a wide variety of biomedical vocabularies have been linked. In this way the UMLS facilitates the mapping of terms between and among any of the components vocabularies that have been linked to the UMLS.
In the field of biomedical information retrieval, two very broad approaches are text-based retrieval and concept-based (or keyword-based) retrieval. Text-based retrieval allows the user to type in arbitrary words or phrases to initiate a search. Google and PubMed are examples of this approach. Concept-based (or keyword-based) retrieval requires that the user provide search terms selected from a restricted vocabulary of concepts (or keywords). Using MeSH terms to search the biomedical literature is one example of this approach. In practice, retrieval systems may allow a combination of concepts (or keywords) and free text to be used. For example, the NLM’s Medline interface allows MeSH terms to be combined with text words in formulating a search of the biomedical literature.
The current pilot NIF CBQI uses the NIFSTD standardized ontology of concepts to construct searches. The NIFSTD ontology is derived in large part from BIRNLex, which was developed for use by the Biomedical Informatics Research Network (BIRN, http://www.nbirn.net/).
The Pilot NIF CBQI: an Example Search
This section uses a simple example to illustrate the operation and capabilities of the current pilot NIF CBQI. The example also helps illustrate concretely some of the design challenges that must be confronted in building such an interface to interact with the three very different NIF resources. In this simple example, the user is interested in information related to purkinje neurons.
Figure 2 shows how a search is formulated. The interface has four components, reflecting the four major steps involved in formulating a search. The first step is labeled “Search for Keywords.” Here the user has entered the text term “purkinje” for this simple example search. After entering this term, the user clicks on the “Search for Keywords” button. This results in a search of the NIFSTD ontology for any concepts (keywords) that match the text word “purkinje.” A list of the concepts found is then displayed in the box labeled “Select Keywords.” In this case three concepts are displayed. The user may then highlight one or more of those concepts and click “Select.” The selected keywords are then copied into the “Compose Query” box.
The main CBQI search page contains four boxes. The first box is used to retrieve keywords (concepts) from the NIFSTD ontology. The second box is used to display those keywords, and to select keywords to be copied to the third box, where the final query is composed. Terms in the “Compose Query” box can be joined using Boolean operators. In the fourth box, the search can be directed to any or all of the NIF resources
The user is able to repeat this process (entering text words or phrases and searching for keywords) several times until he has found and selected a set of one or more concepts that he is satisfied with. Once the desired concepts have been copied into the “Compose Query” box, the user then indicates (using the checkboxes in front of each concept name) which of those concepts he wishes to use in the search. In this simple example, only a single concept is displayed (Purkinje neuron), but in a more complex example two or more concepts might be combined in formulating a search. If several keywords are selected by check boxes, these can be combined using either OR or AND.
The user then specifies (in the fourth box labeled “Retrieve Information”) which of the three NIF resources he wishes to search. In this case, all three resources are checked so all three will be searched. The search is launched by clicking on the “Search” button at the bottom of that part of the screen.
Figure 3 shows how the results of the search are returned to the user. Notice that the page has three Tabs, one Tab for each of the three resources searched. In Fig. 3, the Tab for the NIF Resource Registry is open, so those are the results displayed. Several resources are listed as potentially having information about purkinje neurons. Clicking on the “NIF Entry” link for a resource takes you to a description of that resource in the NIF registry (see Fig. 4). This provides summary information about the resource. Clicking on the “Resource” link, takes the user directly to the Web page for the resource itself (see Fig. 5), from which the user can launch queries directly to the resource, using the “native” Web interface of that resource.
Figure 6 shows the search results from the NIF Document Archive. These results are produced by the Textpresso text-search engine (Müller et al. 2004, 2008). The Textpresso search results in a list of potentially relevant literature citations. Clicking on the “PubMed Link” takes the user directly to the PubMed entry for the paper (see Fig. 7). Alternatively, if the user wants more detailed summary information about each of the citation matches, he can request either 1 or 5 matching sentences for each of the paper (by clicking on “1” or “5” in the sentence just above the results). This request takes the user directly to the Textpresso search engine site, which displays example matching sentences from each matching citation with the search terms highlighted (see Fig. 8). From this page, the user may also launch more detailed searches directly to Textpresso if he so desires.
Results from the Document Archive (returned by the Textpresso search engine) show citations related to Purkinje neuron. From these results, clicking on a “PubMed Link” leads directly to the PubMed citation (Fig. 7). Alternatively the user can request that Textpresso show “1” or “5” matching sentences which transfers the user to Textpresso for Neuroscience for more detail and, if desired, to allow advanced searches related to this query (Fig. 8)
Figure 9 shows how search results are displayed for the NIF Database Mediator. This screen displays different databases that contain potentially relevant data and allows the user to launch a search directly into any one of those databases to retrieve that data. From left to right, we see the names of (1) the database, (2) a table in that database, and (3) fields within that table which may contain relevant information. Each table may have up to two buttons, one (a “Web link out” button labeled with the name of the database) that links to a specific page for the search concept (in this case “Purkinje neuron”) in the resource, and another (“Retrieve Data”) that retrieves information directly from the resource’s back-end database. Note that the search term “Purkinje neuron” has been translated to its corresponding term in each database: e.g., Purkinje neuron (in CCDB - Cell Centered Database (Martone et al. 2008)), and Cerebellar purkinje cell (in SenseLab). Database term translations are performed via the NIF Mediator using mappings between those terms and concepts in the NIFSTD ontology.
This figure shows the initial results from the NIF Database Mediator which displays a list of database tables and fields, containing data about “Purkinje neuron.” From left to right, one sees the database (resource), database table, and the fields within each tables. Terms displayed use the local database terminology. Clicking on the “Retrieve Data” button sends a query to the database to retrieve the data requested via the field’s checkboxes (Fig. 10). Clicking on the “NeuronDB” button transfers the user directly to the dynamically created NeuronDB page containing data about the Cerebellar Purkinje Cell (Fig. 11). Fields containing ‘purkinje neuron’ values are checked and grayed-out because they are automatically used in this query. The presence of a button following a grayed-out field (e.g., NeuronDB) identifies a direct Web link to a page in that resource containing information relevant to the term. Database term translations are performed via the NIF Mediator using mappings between those terms and concepts in the NIFSTD ontology
For each database, the user is given the option of indicating (via checkboxes) which data fields he would like retrieved from each database (by default all fields are selected). For example, for the NeuronDB neuronal current table, if the user clicks on the “Retrieve Data” button he is taken to a new (pop-up) screen (see Fig. 10) containing data about neuronal currents that have been identified in various compartments of the purkinje call. The advantage of this link, is that the data can be inspected in a generic tabular format, and could for example be copied and pasted into a spreadsheet (or into a local database) for integrated analysis with data from other sources.
Data retrieved as requested (see Fig. 9) from the neuronal currents table from NeuronDB
Alternatively, clicking on the “Web Linkout” button (labeled “NeuronDB”) takes the user to the dynamically created Web page in NeuronDB which displays data about the Purkinje call (see Fig. 11). (Such links may not always be available, depending on whether the database is designed to produce that specific page.) The potential advantage of this link is that the user can explore this data using NeuronDB’s native interface that contains a number of capabilities specifically designed to help the user explore and understand this specific type of data, and to compare and relate it to similar data in other neurons.
This figure shows the native NeuronDB Web interface accessed as requested (see Fig. 9) from the NIF Database Mediator
Discussion
The goal of the CBQI is to allow the neuroscientist to compose a single query that can then be run against all three NIF resources. This goal results in a number of challenges, reflecting the very different nature of the three resources. In this section, we discuss certain lessons learned the in the process of designing the CBQI to meet these challenges.
It is worth first emphasizing that the current pilot CBQI has been designed to explore aspects of the concept-based approach. Our goal has not been to make the interface as user-friendly and “seamless” as possible. A free text search interface (such as Google’s) is very easy and intuitive to use. A concept-based approach will need to be more complex, but an important issue for the future will involve exploring how such an interface can be made as intuitive and easy-to-use as possible. In addition, as discussed below, concept-based and free-text searching are potentially synergistic and can likely be productively combined.
The Full Power of the Concept-Based Approach will Only be Achieved when the Database Mediator is Robustly Populated
It is important to emphasize that the most critical need for a concept-based approach to querying the NIF arises because of the Database Mediator. There are many databases available that contain diverse data about the neurosciences. These databases have been built by different research groups and frequently use different, sometimes idiosyncratic, terms and vocabularies.
If the NIF Mediator is to retrieve data from a broad set of these databases, it is essential that any query be formulated in a standardized format with standardized keywords or concepts (e.g., using NIFSTD), and that all the relevant terms in each mediated database be mapped to those terms (as illustrated in Fig. 12). The process of mapping all the relevant terms in a database to the equivalent concepts in NIFSTD is a tedious, time-consuming task. The task is made even more complex by the fact that certain terms in a database may not map to NIFSTD in a one-to-one fashion, due to differences in definitions, differences in granularity (level of detail) of the terms used (e.g., calcium channel vs. different types of calcium channel), etc.
As a result, the expansion of the NIF Database Mediator will be slow compared to the population of the other two NIF resources. Thus the full power of the concept-based approach can only be achieved incrementally over a relatively extended period of time. The Mediator is currently interfaced to five neuroscience databases: NeuronDB, ModelDB, CCDB, Neuromorpho.org, and SumsDB, although only a portion of the information in these databases (approximately 20%) has been mapped to the NIFSTD ontology.
Robust Query of the NIF Resource Registry and Document Archive will Likely Benefit from Combining Concept-based and Textual Retrieval
There are a number of potential problems that arise when applying the concept-based approach to the NIF Resource Registry and to the NIF Document Archive. In the NIF Registry, as mentioned previously, resources are indexed at a quite high level of abstraction. Thus, for example, resources containing data about neurons are indexed with the concept “Neuron.” As a result, if a user has entered the concept “Purkinje neuron”, a number of the resources returned might have data about other types of neurons (e.g., olfactory mitral cells), but not purkinje neurons. In addition, many resources may have data potentially relevant to a concept, but not be indexed by that concept if the relationship is in some was implicit or indirect. As a result, in searching the NIF Registry, it might very well be useful to perform a text search, in addition to the concept-based search, not just of the textual description of the resource in the registry, but also of the Web pages of the resource itself.
The Textpresso search engine is specifically designed to accept textual or conceptual queries. The conceptual queries rely on indexing sentences according to concept names in an ontology. More extensive mappings between the NIFSTD vocabulary and Textpresso concepts, as well as the creation of additional Textpresso concepts, will allow us to take advantage of Textpresso’s conceptual query capability more fully, thereby enhancing its value to the neuroscience user.
As a result of considerations such as these, exploring a query approach that combines a concept-based approach with a text-based approach is a logical future direction. How best to combine the two approaches is far from clear. It does seem clear, however, that a combined approach will likely enhance the ability of the NIF to serve the needs of the neuroscience community.
Extending the Coverage of the NIFSTD Ontology will be Key to Making the Concept-Based Approach Successful
Concept-based querying will only succeed if the ontology of concepts is as comprehensive as possible, and covers most if not all of the concepts of potential interest to neuroscience users. The challenge in accomplishing this goal includes the breadth and diversity of the neuroscience domain and its many intersections with other domains within biomedicine.
In addition, the best approach to developing an ontology for many of the areas within the neurosciences requires much more than a single ontology-builder working in isolation. This task may often require developing a consensus among experts in the field, which is typically a laborious and expensive process. Another complication is that the best ontology for sub-domains within the neurosciences is likely to evolve over time as the scientific field progresses, as the neuroscience phenomena being described become better understood, and as new phenomena are discovered. As a result of all these considerations, a superb ontology for the NIF can only be approached incrementally over time, and will need to undergo a process of regular curation and revision.
As discussed above, the primary need for the concept-based approach is for the Mediator. Since by definition NIFSTD will be linked to the mediated databases, it makes sense to envision an approach where the expansion of NIFSTD is driven in part by the expansion of the databases covered by the NIF Mediator. A mixture of concept-based and text-based search could complement the incremental expansion of NIFSTD by providing broader search capability to all areas of the neuroscience.
A Range of Interesting Issues will Arise due to Ontology Mismatch Among Neuroscience Databases
One issue that will arise in applying the concept-based query approach to the NIF Database Mediator, is that there are bound to be examples of ontology mismatch between the many local database ontologies and the concepts in NIFSTD. Some of these mismatches may reflect a different conceptualization of the neuroscience domain by different research groups and/or an evolving conceptualization that changes over time (for example, in NeuronDB two new oblique dendrite compartments have recently been added to the distal dendrite. Previously, these new oblique dendrite compartments were part of apical dendritic compartments). Other mismatches may reflect the fact that different databases collect data at different levels of detail or in different ways (for example, NeuronDB has neuronal properties assigned to specific neuronal canonical compartments, while Neurodatabase.org (Gardner 2004) uses the approximate distance from the soma when recording specific dendritic properties).
Such ontology mismatches create challenges when trying to help the neuroscientist find and access available data in different databases. Such mismatches will be particularly challenging in the future if the NIF tries to return results from multiple databases in an integrated fashion. The question of how best to deal with ontology mismatches in a complex query system like the NIF presents a major, interesting set of informatics research directions for the future.
The NIF CBQI and the Semantic Web
There is an evolving national initiative that is exploring the use of semantic web technology in the life sciences as a whole, and also specifically within the neurosciences (Lam et al. 2006, 2007; Ruttenberg et al. 2007). Semantic web approaches require that the underlying bioscience concepts be represented using ontologies. This work explores issues such as how ontologies developed for related bioscience domains might best be combined so that data from those domains could be queried in an increasingly integrated fashion. It also explores how additional types of semantic knowledge (e.g., about interrelationships among the concepts) might be included to facilitate more powerful, flexible integration and querying of the data.
Developing a concept-based approach to indexing and querying the NIF represents a major step towards allowing the integration of NIF resources with future efforts to extend and refine the semantic web within the neurosciences and within the life sciences as a whole.
Summary
The present pilot NIF CBQI is allowing us to explore the challenges implicit in applying the concept-based query approach to the diverse and complex domain of the neurosciences. It is also allowing us to explore how best to combine the concept-based and text-based querying approaches. It is clear that particularly as more and more neuroscience databases are incorporated into the NIF Database Mediator, the concept-based approach will provide an essential, powerful tool.
Information Sharing Statement
The CBQI program code is freely available. Please contact the first author.
References
Bug, W., Ascoli, G. A., Grethe, J. S., Gupta, A., Fennema-Notestine, C., Laird, A., et al. (2008). The NIFSTD and BIRNLex vocabularies: Building comprehensive ontologies for neuroscience. Neuroinformatics, doi:10.1007/s12021-008-9032-z.
Gardner, D. (2004). Neurodatabase.org: networking the microelectrode. Nature Neuroscience, 7, 486–487. doi:10.1038/nn0504-486.
Gardner, D., Akil, H., Ascoli, G. A., Bowden, D. M., Bug, W., Donohue, D. E., et al. (2008). The Neuroscience Information Framework: a data and knowledge environment for neuroscience. Neuroinformatics. doi:10.1007/s12021-008-9024-z.
Gupta, A., Bug, W., Marenco, L., Qian, X., Condit, C., Rangarajan, A., et al. (2008). Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF). Neuroinformatics, doi:10.1007/s12021-008-9033-y.
Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research, 32, D258–D261. doi:10.1093/nar/gkh066.
Lam, H. Y., Marenco, L., Clark, T., Gao, Y., Kinoshita, J., Shepherd, G., et al. (2007). AlzPharm: integration of neurodegeneration data using RDF. BMC Bioinformatics, 8(Suppl 3), S4. doi:10.1186/1471-2105-8-S3-S4.
Lam, H. Y., Marenco, L., Shepherd, G. M., Miller, P. L., & Cheung, K. H. (2006). Using web ontology language to integrate heterogeneous databases in the neurosciences. AMIA Symposium, 2006, 464–468.
Martone, M. E., Tran, J., Wong, W. W., Sargis, J., Fong, L., Larson, S., et al. (2008). The cell centered database project: an update on building community resources for managing and sharing 3D imaging data. Journal of Structural Biology, 161, 220–231. doi:10.1016/j.jsb.2007.10.003.
Müller, H. M., Kenny, E. E., & Sternberg, P. W. (2004). Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biology, 2, e309. doi:10.1371/journal.pbio.0020309.
Müller, H. M., Rangarajan, A., Teal, T. K., & Sternberg, P. W. (2008). Textpresso for neuroscience: searching the full text of thousands of neuroscience research papers. Neuroinformatics. doi:10.1007/s12021-008-9031-0.
Ruttenberg, A., Clark, T., Bug, W., Samwald, M., Bodenreider, O., Chen, H., et al. (2007). Advancing translational research with the Semantic Web. BMC Bioinformatics, 8(Suppl 3), S2. doi:10.1186/1471-2105-8-S3-S2.
Acknowledgments
This project has been funded in whole or in part through the NIH Blueprint for Neuroscience Research with Federal funds from the National Institute on Drug Abuse, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN271200577531C. This research was also supported by
• NIH grants P01 DC04732 and R01 DA021253,
• Volunteer consultant-collaborators and friends, and
• The Society for Neuroscience.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Marenco, L., Li, Y., Martone, M.E. et al. Issues in the Design of a Pilot Concept-Based Query Interface for the Neuroinformatics Information Framework. Neuroinform 6, 229–239 (2008). https://doi.org/10.1007/s12021-008-9035-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-008-9035-9