Abstract
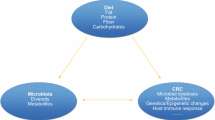
Colorectal cancer (CRC) is the most common gastrointestinal cancer, and a significant health care problem globally. Dietary factors, for example high meat consumption and deficiency of fiber, calcium, vitamin D, and folate, are well-recognized to be associated with a risk of developing CRC. Colonic microbiota, by living in a mutual relationship and participating in key metabolic functions that compliment host physiology, is crucially important in the maintenance of our health. A state of imbalance in host–microbe homeostasis, termed dysbiosis, is associated with several diseases, including CRC. Epidemiological studies have revealed strong associations between diet, microbiota, and CRC. Substantial in-vitro and in-vivo evidence suggests that the dynamic composition and diversity of colonic microbiota are affected by alteration of the diet, and that the balance between the beneficial and detrimental microbial metabolites is of crucial importance in mediation of the dietary risk factors of colonic carcinogenesis. A better understanding of complex diet–microbiota–CRC relationships can help us understand how diet affects the risk of CRC and will provide a more scientific approach to the development of novel strategies to prevent CRC.

Similar content being viewed by others
Abbreviations
- AA:
-
African American
- AICR:
-
American Institute for Cancer Research
- AOM:
-
Azoxymethane
- B-12:
-
Cyanocobalamin
- BA:
-
Bile acids
- CA:
-
Caucasian American
- CDA:
-
Chenodeoxycholic acid
- CH4 :
-
Methane
- COX:
-
Cycloxygenase
- DCA:
-
Deoxycholic acid
- EPEC:
-
Entero-pathogenic Escherichia coli
- EPIC:
-
European Prospective Investigation into Cancer and Nutrition
- ETBF:
-
Entero-toxigenic Bacteroides fragilis
- EA:
-
European African
- GF:
-
Germ-free
- H2 :
-
Hydrogen
- H2S:
-
Hydrogen sulfide
- HCA:
-
Heterocylic amines
- IBD:
-
Inflammatory Bowel Disease
- LCA:
-
Lithocholic acid
- MA:
-
Methanogenic archaea
- MALT:
-
Mucosa-Associated Lymphoid Tissue
- NA:
-
Native African
- NOC:
-
N-nitroso compounds
- PAH:
-
Polycyclic aromatic hydrocarbons
- ROS:
-
Reactive oxygen species
- RNS:
-
Reactive nitrogen species
- RS:
-
Resistant Starch
- SCFA:
-
Short-chain fatty acids
- SRB:
-
Sulfate-reducing bacteria
- WCRF:
-
World Cancer Research Fund
References
Papers of particular interest, published recently have been highlighted as: • Of importance
Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst. 1981;66(6):1191–308.
Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol Syst Biol. 2006;2:52.
Wardwell LH, Huttenhower C, Garrett WS. Current concepts of the intestinal microbiota and the pathogenesis of infection. Curr Infect Dis Rep. 2011;13(1):28–34.
Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol. 1998;6(1–2):13–8.
O’Keefe SJ, Winter TA, Newton KA, Ogden JM, Young GO, Price SK. Severe malnutrition associated with alpha-heavy chain disease: response to tetracycline and intensive nutritional support. Am J Gastroenterol. 1988;83(9):995–1001.
Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6(8):229.
• Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. The human microbiome project was launched to study and characterize the ecology of human microbiome, which was found to vary both within and between the individuals, enabling future characterization of the epidemiology, ecology, and translational applications of the human microbiome.
• Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80. In their fecal metagenomics analysis, the authors have, for the first time, identified distinct clustering of the microbiome into three main ‘enterotypes’ independent of age, gender or country, depending on their common networks of co- and anti-correlating genera.
Flood DM, Weiss NS, Cook LS, Emerson JC, Schwartz SM, Potter JD. Colorectal cancer incidence in Asian migrants to the United States and their descendants. Cancer Causes Control. 2000;11(5):403–11.
Gonzalez CA. The European Prospective Investigation into Cancer and Nutrition (EPIC). Public Health Nutr. 2006;9(1A):124–6.
• Cross AJ, Ferrucci LM, Risch A, et al. A large prospective study of meat consumption and colorectal cancer risk: an investigation of potential mechanisms underlying this association. Cancer Res. 2010;70(6):2406–14. In this very large US prospective study of 2,719 CRC cases in a cohort of 300,948 men and women, the authors found a positive association of red and processed meat intake with CRC and suggested involvement of heme iron, nitrate/nitrite, and HCA from meat as a possible explanation of these associations.
Culp SJ, Gaylor DW, Sheldon WG, Goldstein LS, Beland FA. A comparison of the tumors induced by coal tar and benzo[a]pyrene in a 2-year bioassay. Carcinogenesis. 1998;19(1):117–24.
Hughes R, Magee EA, Bingham S. Protein degradation in the large intestine: relevance to colorectal cancer. Curr Issues Intest Microbiol. 2000;1(2):51–8.
de Vogel J, Van-Eck WB, Sesink AL, Jonker-Termont DS, Kleibeuker J, van der Meer R. Dietary heme injures surface epithelium resulting in hyperproliferation, inhibition of apoptosis and crypt hyperplasia in rat colon. Carcinogenesis. 2008;29(2):398–403.
Mirvish SS. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 1995;93(1):17–48.
Cross AJ, Pollock JR, Bingham SA. Haem, not protein or inorganic iron, is responsible for endogenous intestinal N-nitrosation arising from red meat. Cancer Res. 2003;63(10):2358–60.
Ward MH, Cross AJ, Divan H, et al. Processed meat intake, CYP2A6 activity and risk of colorectal adenoma. Carcinogenesis. 2007;28(6):1210–6.
Bingham SA, Hughes R, Cross AJ. Effect of white versus red meat on endogenous N-nitrosation in the human colon and further evidence of a dose response. J Nutr. 2002;132(11 Suppl):3522S–5S.
Kazerouni N, Sinha R, Hsu CH, Greenberg A, Rothman N. Analysis of 200 food items for benzo[a]pyrene and estimation of its intake in an epidemiologic study. Food Chem Toxicol. 2001;39(5):423–36.
Schut HA, Snyderwine EG. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20(3):353–68.
Hasegawa R, Sano M, Tamano S, et al. Dose-dependence of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) carcinogenicity in rats. Carcinogenesis. 1993;14(12):2553–7.
Kassie F, Rabot S, Kundi M, Chabicovsky M, Qin HM, Knasmuller S. Intestinal microflora plays a crucial role in the genotoxicity of the cooked food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline. Carcinogenesis. 2001;22(10):1721–5.
• Moschen AR, Wieser V, Tilg H. Dietary factors: major regulators of the Gut’s microbiota. Gut Liver. 2012;6(4):411–6. In this up-to-date review, the authors summarized the effects of different diets on human microbiome composition and diversity.
Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–24.
Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14.
• De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–6. In their study of the effect of diet on microbiota, the authors noted significant differences between the fecal microbiomes of European and African children and implicated their different diets (western vs African diet) as the reason.
• Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–8. In their analysis of the diet inventories and fecal microbiomes of subjects from four countries, the authors noted these correlated with their diet in the long term, supporting the involvement of the diet in the modulation of microbiota.
Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5(5):455–9.
Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–59.
• Conlon MA, Kerr CA, McSweeney CS, et al. Resistant starches protect against colonic DNA damage and alter microbiota and gene expression in rats fed a Western diet. J Nutr. 2012;142(5):832–40. By supplementing rats fed on a western diet with resistant starch that enhanced SCFA and minimized ammonia and phenol levels, the authors noted that resistant starch opposed western diet-induced colonic DNA damage.
Dolara P, Luceri C, De Filippo C, et al. Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat Res. 2005;591(1–2):237–46.
• Devkota S, Wang Y, Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487(7405):104–8. In this study the authors have noted that milk-derived fat stimulated taurine rich bile acids which promoted the growth of a sulfite (in taurine)-reducing B. wadsworthia, that was associated with colitis in Il10 −/− mice, offering a possible mechanism of western diet-related gastrointestinal diseases.
Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immunol. 1998;66(11):5224–31.
Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4(6):0006026.
Swidsinski A, Khilkin M, Kerjaschki D, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115(2):281–6.
Abdulamir AS, Hafidh RR, Abu Bakar F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exp Clin Cancer Res. 2011;30(11):1756–9966.
Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1(3):138–47.
Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1):0016393.
Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306.
Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One. 2011;6(5):24.
Scanlan PD, Shanahan F, Clune Y, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. 2008;10(3):789–98.
O’Keefe SJ, Chung D, Mahmoud N, et al. Why do African Americans get more colon cancer than Native Africans? J Nutr. 2007;137(1 Suppl):175S–82S.
O’Keefe SJ, Ou J, Aufreiter S, et al. Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J Nutr. 2009;139(11):2044–8.
Fukata M, Chen A, Vamadevan AS, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133(6):1869–81.
• Wu WK, Sung JJ, Lee CW, Yu J, Cho CH. Cyclooxygenase-2 in tumorigenesis of gastrointestinal cancers: an update on the molecular mechanisms. Cancer Lett. 2010;295(1):7–16. In this elaborate review the authors explain how NSAIDs suppress gastrointestinal tumors on the basis of the involvement of COX-2, PG-E2, and the signaling mechanisms involved.
Humblot C, Murkovic M, Rigottier-Gois L, et al. Beta-glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in rats. Carcinogenesis. 2007;28(11):2419–25.
Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. 2008;66(3):487–95.
Kim DH, Jin YH. Intestinal bacterial beta-glucuronidase activity of patients with colon cancer. Arch Pharm Res. 2001;24(6):564–7.
Hijova E, Bomba A, Bertkova I, Strojny L, Szabadosova V, Soltesova A. Prebiotics and bioactive natural substances induce changes of composition and metabolic activities of the colonic microflora in cancerous rats. Acta Biochim Pol. 2012;59(2):271–4.
Gorbach SL, Goldin BR. The intestinal microflora and the colon cancer connection. Rev Infect Dis. 1990;12(2):S252–61.
Roldan MD, Perez-Reinado E, Castillo F, Moreno-Vivian C. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol Rev. 2008;32(3):474–500.
Nakamura J, Kubota Y, Miyaoka M, Saitoh T, Mizuno F, Benno Y. Comparison of four microbial enzymes in Clostridia and Bacteroides isolated from human feces. Microbiol Immunol. 2002;46(7):487–90.
Cummings JH, Englyst HN. Measurement of starch fermentation in the human large intestine. Can J Physiol Pharmacol. 1991;69(1):121–9.
Chirakkal H, Leech SH, Brookes KE, Prais AL, Waby JS, Corfe BM. Upregulation of BAK by butyrate in the colon is associated with increased Sp3 binding. Oncogene. 2006;25(54):7192–200.
Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132(5):1012–7.
Comalada M, Bailon E, de Haro O, et al. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol. 2006;132(8):487–97.
Andoh A, Shimada M, Araki Y, Fujiyama Y, Bamba T. Sodium butyrate enhances complement-mediated cell injury via down-regulation of decay-accelerating factor expression in colonic cancer cells. Cancer Immunol Immunother: CII. 2002;50(12):663–72.
Zgouras D, Wachtershauser A, Frings D, Stein J. Butyrate impairs intestinal tumor cell-induced angiogenesis by inhibiting HIF-1alpha nuclear translocation. Biochem Biophys Res Commun. 2003;300(4):832–8.
Zeng H, Briske-Anderson M. Prolonged butyrate treatment inhibits the migration and invasion potential of HT1080 tumor cells. J Nutr. 2005;135(2):291–5.
Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(7 Suppl):2485S–93S.
Place RF, Noonan EJ, Giardina C. HDAC inhibition prevents NF-kappa B activation by suppressing proteasome activity: down-regulation of proteasome subunit expression stabilizes I kappa B alpha. Biochem Pharmacol. 2005;70(3):394–406.
Schwab M, Reynders V, Ulrich S, Zahn N, Stein J, Schroder O. PPARgamma is a key target of butyrate-induced caspase-3 activation in the colorectal cancer cell line Caco-2. Apoptosis: Int J Program Cell Death. 2006;11(10):1801–11.
Karaki S, Tazoe H, Hayashi H, et al. Expression of the short-chain fatty acid receptor, GPR43, in the human colon. J Mol Histol. 2008;39(2):135–42.
Brown AJ, Goldsworthy SM, Barnes AA, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278(13):11312–9.
Willemsen LE, Koetsier MA, van Deventer SJ, van Tol EA. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52(10):1442–7.
Schauber J, Svanholm C, Termen S, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52(5):735–41.
Malago JJ, Koninkx JF, Tooten PC, van Liere EA, van Dijk JE. Anti-inflammatory properties of heat shock protein 70 and butyrate on Salmonella-induced interleukin-8 secretion in enterocyte-like Caco-2 cells. Clin Exp Immunol. 2005;141(1):62–71.
Puchowicz MA, Bederman IR, Comte B, et al. Zonation of acetate labeling across the liver: implications for studies of lipogenesis by MIDA. Am J Physiol. 1999;277(6 Pt 1):E1022–7.
Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. 2010;1801(11):1175–83.
Elsden SR, Hilton MG, Waller JM. The end products of the metabolism of aromatic amino acids by Clostridia. Arch Microbiol. 1976;107(3):283–8.
Russell JB. Fermentation of Peptides by Bacteroides ruminicola B(1)4. Appl Environ Microbiol. 1983;45(5):1566–74.
Cummings JH, Hill MJ, Bone ES, Branch WJ, Jenkins DJ. The effect of meat protein and dietary fiber on colonic function and metabolism. II. Bacterial metabolites in feces and urine. Am J Clin Nutr. 1979;32(10):2094–101.
Nowak A, Libudzisz Z. Ability of probiotic Lactobacillus casei DN 114001 to bind or/and metabolise heterocyclic aromatic amines in vitro. Eur J Nutr. 2009;48(7):419–27.
Reddy BS. Diet and excretion of bile acids. Cancer Res. 1981;41(9 Pt 2):3766–8.
McGarr SE, Ridlon JM, Hylemon PB. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol. 2005;39(2):98–109.
Haines A, Hill MJ, Thompson MH, et al. A prospective study of faecal bile acids and colorectal cancer. Eur J Cancer Prev. 2000;9(5):317–23.
• Ou J, DeLany JP, Zhang M, Sharma S, O’Keefe SJ. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr Cancer. 2012;64(1):34–40. The authors noted that high fecal bile acids (with carcinogenic properties) and low SCFA (with anti-inflammatory and anti-neoplastic properties) explain the high colon cancer risk of a high red meat and low fiber diet, emphasizing the involvement of microbiota and metabolites in mediation of diet-related CRC risk.
Cheng K, Raufman JP. Bile acid-induced proliferation of a human colon cancer cell line is mediated by transactivation of epidermal growth factor receptors. Biochem Pharmacol. 2005;70(7):1035–47.
Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. 2009;15(27):3329–40.
Powolny A, Xu J, Loo G. Deoxycholate induces DNA damage and apoptosis in human colon epithelial cells expressing either mutant or wild-type p53. Int J Biochem Cell Biol. 2001;33(2):193–203.
Booth LA, Gilmore IT, Bilton RF. Secondary bile acid induced DNA damage in HT29 cells: are free radicals involved? Free Radic Res. 1997;26(2):135–44.
Qiao D, Gaitonde SV, Qi W, Martinez JD. Deoxycholic acid suppresses p53 by stimulating proteasome-mediated p53 protein degradation. Carcinogenesis. 2001;22(6):957–64.
Jurek D, Fleckl E, Marian B. Bile acid induced gene expression in LT97 colonic adenoma cells. Food Chem Toxicol. 2005;43(1):87–93.
Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15(5):2156–63.
Gibson GR, Macfarlane GT, Cummings JH. Sulphate reducing bacteria and hydrogen metabolism in the human large intestine. Gut. 1993;34(4):437–9.
Wolin M. Microbial formation and utilization of gases. Gottingen: Goltze Press; 1976.
Gibson GR, Cummings JH, Macfarlane GT. Competition for hydrogen between sulphate-reducing bacteria and methanogenic bacteria from the human large intestine. J Appl Bacteriol. 1988;65(3):241–7.
O’Keefe SJ, Kidd M, Espitalier-Noel G, Owira P. Rarity of colon cancer in Africans is associated with low animal product consumption, not fiber. Am J Gastroenterol. 1999;94(5):1373–80.
Christl SU, Eisner HD, Dusel G, Kasper H, Scheppach W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: a potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci. 1996;41(12):2477–81.
Kanazawa K, Konishi F, Mitsuoka T, et al. Factors influencing the development of sigmoid colon cancer. Bacteriologic and biochemical studies. Cancer. 1996;77(8 Suppl):1701–6.
Ramasamy S, Singh S, Taniere P, Langman MJ, Eggo MC. Sulfide-detoxifying enzymes in the human colon are decreased in cancer and upregulated in differentiation. Am J Physiol Gastrointest Liver Physiol. 2006;291(2):23.
• Nava GM, Carbonero F, Ou J, Benefiel AC, O’Keefe SJ, Gaskins HR. Hydrogenotrophic microbiota distinguish native Africans from African and European Americans. Environ Microbiol Rep. 2012;4:307–15. The authors noted that high dietary resistant starch/fiber promotes the abundance of colonic methanogens (which detoxify H 2 into benign methane) whereas a high red meat diet promotes SRB (which convert H 2 into toxic H 2 S), emphasizing that diet selects for hydrogenic microbiota.
Pompei A, Cordisco L, Amaretti A, et al. Administration of folate-producing bifidobacteria enhances folate status in Wistar rats. J Nutr. 2007;137(12):2742–6.
Kim YI. Role of folate in colon cancer development and progression. J Nutr. 2003;133(11 Suppl 1):3731S–9S.
• Acharya A, Das I, Chandhok D, Saha T. Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxid Med Cell Longev. 2010;3(1):23–34. In this review the authors describe how oxidative stress and reactive oxygen species cause DNA damage and carcinogenesis.
Huycke MM, Joyce W, Wack MF. Augmented production of extracellular superoxide by blood isolates of Enterococcus faecalis. J Infect Dis. 1996;173(3):743–6.
Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23(3):529–36.
Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132(2):551–61.
Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23(8 Pt 1):1298–303.
Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124(2):392–400.
Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–22.
Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107(25):11537–42.
Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4(5):13.
Disclosure
No potential conflicts of interest relevant to this article were reported.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vipperla, K., O’Keefe, S.J. Intestinal Microbes, Diet, and Colorectal Cancer. Curr Colorectal Cancer Rep 9, 95–105 (2013). https://doi.org/10.1007/s11888-012-0158-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11888-012-0158-x