Abstract
Many important cellular processes are regulated by reaction-diffusion (RD) of molecules that takes place both in the cytoplasm and on the membrane. To model and analyze such multicompartmental processes, we developed a lattice-based Monte Carlo method, Spatiocyte that supports RD in volume and surface compartments at single molecule resolution. Stochasticity in RD and the excluded volume effect brought by intracellular molecular crowding, both of which can significantly affect RD and thus, cellular processes, are also supported. We verified the method by comparing simulation results of diffusion, irreversible and reversible reactions with the predicted analytical and best available numerical solutions. Moreover, to directly compare the localization patterns of molecules in fluorescence microscopy images with simulation, we devised a visualization method that mimics the microphotography process by showing the trajectory of simulated molecules averaged according to the camera exposure time. In the rod-shaped bacterium Escherichia coli, the division site is suppressed at the cell poles by periodic pole-to-pole oscillations of the Min proteins (MinC, MinD and MinE) arising from carefully orchestrated RD in both cytoplasm and membrane compartments. Using Spatiocyte we could model and reproduce the in vivo MinDE localization dynamics by accounting for the previously reported properties of MinE. Our results suggest that the MinE ring, which is essential in preventing polar septation, is largely composed of MinE that is transiently attached to the membrane independently after recruited by MinD. Overall, Spatiocyte allows simulation and visualization of complex spatial and reaction-diffusion mediated cellular processes in volumes and surfaces. As we showed, it can potentially provide mechanistic insights otherwise difficult to obtain experimentally.
Similar content being viewed by others
Introduction
Computational models are valuable to both theoretical and experimental biologists to rapidly analyze and test their predictions about qualitative or quantitative behavior of complex cellular processes (Ideker and Lauffenburger 2003). Moreover, the models can potentially provide mechanistic insights (Treanor and Batista 2007; Clarke and Liu 2008; Neves and Iyengar 2009), even in the case of relatively simple processes (Stefan et al. 2008; van Albada and ten Wolde 2009).
Processes that are strongly dependent on the cell morphology, multicompartmental interaction and dynamic localization of molecules such as cell signaling (Rangamani and Iyengar 2008; Neves et al. 2008), bacterial cell division (Lutkenhaus 2007; Adams and Errington 2009) and chemotaxis (Greenfield et al. 2009; Rao and Ordal 2009), require realistic models with spatial representation (Takahashi et al. 2005; Ridgway et al. 2006; Morris and Jensen 2008). The processes rely on intricately controlled reaction and diffusion of molecules in and between cytoplasm (or nucleoplasm) and membrane compartments to perform their functions. Excluded volume effect brought by molecular crowding in each compartment can reduce the macroscopic diffusion coefficient and alter reaction kinetics (Ellis 2001; Hall and Minton 2003; Dix and Verkman 2008; Klann et al. 2009). Furthermore, the significance of slower diffusion in such reaction-diffusion (RD) processes is pronounced when the reactions are diffusion-limited. Since some of the species involved in the processes comprise small number of molecules, stochasticity in the reactions can dominate over the law of mass action (Gillespie 1976; Gómez-Uribe and Verghese 2007), and thus affecting the macroscopic behavior of the processes (Fange and Elf 2006; Rangamani and Iyengar 2008; Hansen et al. 2008).
The prominent spatial modeling methods are listed in Table 1 with a summary of their features. The table shows that although a variety of methods have been developed, modeling cellular processes at the level where multicompartmental RD, molecular crowding and stochasticity all simultaneously come into play is still not possible with any single method. To address this issue, we developed a general-purpose hybrid time- and event-driven, lattice-based Monte Carlo simulation method, called Spatiocyte, to model processes that are dependent on multicompartmental RD, stochasticity and the implications of molecular crowding.
In the second part of this study, using Spatiocyte we modeled the Escherichia coli MinDE division site control system and investigate the mechanism of MinE ring formation, which remains unclear (Lutkenhaus 2007). The rod-shaped bacterium begins to divide when the tubulin-homolog FtsZ polymerizes into a constricting annular structure (Z-ring) on the inner membrane (Adams and Errington 2009; Lutkenhaus 2007). To ensure equal distribution of cell contents to the two progeny cells, the constriction site is restricted to the middle of the cell long axis by two mechanisms: (i) nucleoid occlusion, which allows the Z-ring to assemble only at the midcell and the cell poles, locations that are devoid of the nucleoid mass (Woldringh et al. 1990; 1991; Bernhardt and de Boer 2005), and (ii) carefully orchestrated RD of the minB operon encoded proteins, MinC, MinD, and MinE, in the cytoplasm and inner membrane, which suppresses polar Z-rings to avoid the minicelling phenotype (Lutkenhaus 2007; Vats et al. 2009). The phenotype is characterized by the formation of chromosomeless minicells and elongated cells with multiple chromosomes via polar septation.
In the absence of MinE, the ATPase MinD in the ATP form, MinDATP binds peripherally around the entire membrane (Raskin and de Boer 1999). MinE, however, induces the MinD ATPase activity on the membrane, causing hydrolyzed MinDADP to be released into the cytoplasm and membrane-associated MinDATP to localize into a polar zone that oscillates between the poles with a period of approximately 1 min (Hu and Lutkenhaus 2001). The FtsZ inhibitor, MinC piggybacks on the oscillating MinDATP (Hu et al. 2003; Zhou and Lutkenhaus 2004), ensuring that its time-averaged concentration is lowest at the midcell for the Z-ring assembly while sufficiently high at the poles to abrogate nascent FtsZ polymers (Hu et al. 1999; Hu and Lutkenhaus 1999).
While the exact mechanism is not known, MinE also localizes annularly to form the E-ring at the medial edge of the polar zone (Raskin and de Boer 1997). When the E-ring is disrupted, the polar zone extends over the midcell and oscillates markedly slower, resulting in the minicelling phenotype (Raskin and de Boer 1997; Zhang et al. 1998; Shih et al. 2002). In the absence of MinD, wild type MinE is always cytosolic. However, based on the observations from recent studies we predict that the MinE constituting the E-ring could be transiently attached to the membrane without being directly tethered to membrane-bound MinD. Firstly, double-label fluorescence microscopy experiments by Shih et al. (2002) clearly indicate that there is a higher density of MinE in the wild type E-ring than that of MinD at the rim of the polar zone, ruling out the possibility that all membrane-recruited MinE subunits are MinD-bound. In addition, the width of the observed E-ring, which appears to be several times the length of a MinE dimer, is also unlikely to be the direct result of MinE dimers binding to MinD subunits at the rim. Truncated MinE N-terminal domain can autonomously bind to the membrane in the absence of MinD, and in the full length MinE, the binding domain is thought to be exposed upon membrane recruitment by MinD (Ma et al. 2003).
Even though many computational models of the MinDE system have been proposed (Howard et al. 2001; Meinhardt and de Boer 2001; Kruse 2002; Howard and Rutenberg 2003; Huang et al. 2003; Drew et al. 2005; Kerr et al. 2006; Fange and Elf 2006; Pavin et al. 2006; Tostevin and Howard 2006; Cytrynbaum and Marshall 2007; Loose et al. 2008; Derr et al. 2009; Borowski and Cytrynbaum 2009), the possibility that MinE is transiently attached to the membrane to form the dense E-ring has not been investigated at single molecule resolution in three-dimensional (3D) space. Here by accounting for the transient membrane attachment of MinE, we modeled the MinDE system with Spatiocyte and successfully reproduced the observed in vivo dynamic localization patterns. Our simulation results suggest that the transient membrane attachment of MinE leads to the formation of E-ring.
Methods
Multicompartmental simulation space
In Spatiocyte, molecules can participate in first- or second-order reactions, each typically giving away one or two product molecules. In vivo, where the compartments are crowded (Zimmerman and Trach 1991; Dupuy and Engelman 2008), diffusing molecules collide even at very small time scales (Ellis 2001). The advantage of completely event-driven continuum RD methods (see Table 1 and Popov and Agmon 2001) to make large jumps in time when molecules are far apart is lost in such conditions. Instead, we use a time-driven, lattice-based method to skip two costly operations—molecule search and distance calculation—required by off-lattice methods to resolve collisions (Ridgway et al. 2006; Takahashi et al. 2005). Diffusion-independent reactions, however, are event driven to further reduce the computational cost.
We discretized the space into an hexagonal close-packed (HCP) lattice because it has the highest density of sphere voxels (Szpiro 2003). Each voxel has 12 adjoining but non-overlapping neighbors (Fig. 1a) and is at most occupied by a single molecule to account for volume exclusion and molecular crowding. The voxel radius r v is uniform and set to the radius of individually simulated molecules. To delineate a surface compartment such as a cell or a nuclear membrane, we occupy all empty voxels of the compartment with immobile lipid molecules (Fig. 1b). Analogous to in vivo, these molecules aid lateral diffusion of non-lipid surface species, while together as an immobile structure, sequester volume species.
Multicompartmental simulation space discretized into hexagonal close-packed (HCP) lattice. a A voxel in the HCP lattice (blue) has 12 adjoining neighbor voxels. b Cross-section of cell (teal) and nuclear (maroon) membranes represented by immobile lipid molecules that constrain molecules to their respective compartment. Other colored spheres represent molecules in their respective voxel. Various geometric primitives such as boxes, spheres and cylinders can be used in combinations to define compartment structures, cell shapes and intracellular geometries, which may affect the modeled system behavior. The rod-shaped structure of Escherichia coli for example, is crucial for the correct oscillation behavior of the MinDE system. The bacterium cell structure can be defined with a cylinder as the body and two hemispheres as the poles, forming the overall inner membrane, while the cytoplasm is represented by the enclosed volume
In addition to the diffusing and immobile species that are treated individually at the molecular scale, there are also homogeneously distributed (HD) species, such as ATP, that are simulated at the compartmental scale. We consider a species HD only if it is mobile and evenly distributed within the compartment before its reaction events. The species can satisfy these conditions if its reaction events are not limited by its diffusion rate. Alternatively, it can have a sufficiently high concentration that, after each reaction event, even if it has a slower diffusion rate it can rapidly attain a homogeneous concentration before the next event. Each molecule of the HD species is not treated individually since the compartmental concentration information is sufficient. For a biochemical species A, we denote A # as the total molecule number in a compartment. All random numbers are drawn independently from the uniform distribution.
Diffusion
Applying the well-known Einstein-Smoluchowski expression for diffusion \(<r^{2}>=2lD_i\tau_i\) , with l = 3 for volume diffusion, a molecule i with diffusion coefficient D i can walk to a randomly selected neighbor voxel in a time step τ i when the mean squared displacement \(<r^{2}>=(2r_v)^2\) (Rodríguez et al. 2006). For a surface molecule, the walk is constrained to adjoining surface voxels, l = 2 and the average voxel-to-voxel lateral displacement is \(\frac{2(2\sqrt{2}+4\sqrt{3}+3\sqrt{6}+\sqrt{22})r_v} {6\sqrt{2}+4\sqrt{3}+3\sqrt{6}}\approx1.915r_v\) (see supporting Fig. S1). Thus, if t s is the current simulation time, the next time a diffusing molecule walks is given by
where \(\alpha_i=\frac{2}{3}\) or \(\alpha_i=\left(\frac{2\sqrt{2}+4\sqrt{3}+3\sqrt{6}+\sqrt{22}} {6\sqrt{2}+4\sqrt{3}+3\sqrt{6}}\right)^2\) for volume or surface species, respectively. However, a volume molecule can only occupy the target voxel if it is vacant, otherwise a collision occurs and the walk fails. A surface walk is only successful if the target voxel is occupied by a lipid molecule, in which case the lipid and the walking molecule exchange voxels; a collision occurs if the target voxel contains a non-lipid surface species. The above scheme accurately constrains molecules to their own compartment without geometry calculations.
Molecules sharing a diffusion coefficient D i are grouped in a diffusion process d i which executes their random walk together at t i d . In long time-scales, diffusion becomes a costly operation because of the very short time steps required to make voxel-length random walks. To alleviate this cost, every voxel has an address pointer to each of its neighbors as a rapid means of access when checking for vacancy and molecule species. In addition, only mobile molecules undergoing diffusion-influenced reactions, usually found in small numbers, are individually diffused, while others are simulated at the compartmental scale as HD species.
Diffusion-independent reactions
\(A\,\xrightarrow{k_A}\,\hbox{product(s)}\) is a first-order reaction with the product(s) comprising diffusing, immobile or HD species. It can represent a unimolecular reaction, which can be decoupled from the diffusion process since k A is only dependent on time. It can also denote a bimolecular reaction (i.e., pseudo first-order), involving A and an unrepresented HD species with a fixed concentration. The reaction is decoupled from diffusion by setting k A as the asymptotic reaction rate given by k a πRD/(k a + 4πRD), where k a is the intrinsic reaction rate, while R and D are the contact distance and combined diffusion coefficient, respectively.
Applying the asymptotic reaction rate, a second-order reaction involving a HD reactant can also be decoupled from the diffusion or immobility of the second reactant because before the reaction event, the concentration of the HD species is the same at any location of the compartment. Likewise, if both reactants are immobile, as long as they are in contact (via adjoining voxels in the simulation), the rate of reaction is also unaffected. Taken together, we can decouple diffusion from all first-order reactions, and all second-order reactions that involve two adjoining immobile reactants or at least one HD reactant.
We perform event-driven diffusion-independent reactions by adapting the Next Reaction (NR) method (Gibson and Bruck 2000), a computer-optimized variation of the Gillespie algorithm (Gillespie 1976; 1977) that stores reaction dependencies in a directed graph and the time of next reactions in an indexed priority queue. The steps in an iteration of the NR method are as follows: (i) select the reaction m with the earliest next reaction time t m r from the priority queue; (ii) execute reaction m and update species molecule numbers; (iii) using the dependency graph, get every reaction μ whose propensity a μ is affected by the updated species and recompute it:
where k S is the surface-average rate coefficient (with dimensions of velocity (Balgi et al. 1995)) for converting the volume species A to surface product(s), S is the surface compartment area, and V is the compartment volume, which is replaced with S if both reactants in the second-order reactions are surface species; and (iv) for each affected reaction, using a random number u r in the range (0,1) update the priority queue with the new next reaction time
The original method assumes all molecules to be HD species, which is not the case in our approach. As a result, in step (ii), if m involves a non-HD reactant species, we perform the reaction with a molecule randomly picked from the non-HD reactant molecules. A non-HD product of m replaces a non-HD reactant at the same voxel. A second non-HD product will occupy a vacant neighbor voxel of the first product. A non-HD product of m involving only HD reactants is placed in a random vacant voxel of the reaction compartment. If it is an intercompartmental reaction, the product will take up a vacant voxel adjoining both compartments.
Diffusion-influenced reactions
Only two remaining second-order reactions, involving a diffusing and an immobile reactant, and two diffusing reactants, are diffusion influenced. They are modelled using a spatially discretized version of the Collins and Kimball approach (Collins and Kimball 1949) that assigns a finite probability p for a reactive collision between reactants.
Consider a diffusion-influenced reaction j between A and B that occurs in the HCP lattice. If A is mobile, its diffusion process will walk each A after the interval \(\tau_A=\frac{\alpha_A{r_v}^2}{D_A}\). A collision with B at the target voxel will set off the reaction with probability p j . In supplementary information, we have derived the relationship between p j and the macroscopic rate constant of various diffusion-influenced second-order reactions:
where \(\gamma=\frac{(2\sqrt{2}+4\sqrt{3}+3\sqrt{6}+\sqrt{22})^2} {72(6\sqrt{2}+4\sqrt{3}+3\sqrt{6})}\). The species subscripts v and s denote volume and surface species respectively, while L is the lipid species.
If j is diffusion-limited, a reactive collision may occur at time slices smaller than τ A , causing p j > 1 and yielding inaccurate results. Here, for a species i that participates in J number of diffusion-influenced reactions, we limit its reaction probabilities to a maximum value P i , 0 ≤ P i ≤ 1, by reducing its diffusion process interval to \(\tau_i'=\tau_iP_i/\rho_i\) , with \(\rho_i=\max\{p_1,\ldots,p_J\}\). Consequently, Eq. 1 becomes
Note that if ρ i > P i , the walk probability is no longer unity because of the reduced interval and is given as P i /ρ i . However, at each interval the molecule can collide as usual with a neighbor reactant pair and react with probability p j P i /ρ i .
When the collision is reactive, the two reactants are removed and new non-HD product(s) occupy the reactant or neighbor voxels, whereas for HD product(s), only the species molecule number is updated. Since the reactant and product molecule numbers have changed, we also perform steps (iii) and (iv) of the adapted NR method for the diffusion-independent reactions.
Hybrid time- and event-driven simulation
We adopt the multi-timescale algorithm of the E-Cell System (Takahashi et al. 2004) to concurrently run time- and event-driven processes. All n number of t i d and t μ r are stored in an indexed priority queue Q, which provides \(t_q=\min\{t^i_d,t^\mu_r\}\) and can be updated in O(logn) (Takahashi et al. 2004). If \(t_q=t^i_d\), d i is executed and Q is updated with a new next walk time from (5). If t q = t μ r , the steps (ii) to (iv) of the adapted NR method are executed using Q with m = μ. By incrementing t s = t s + t q , we can run diffusion processes at predefined time steps while diffusion-independent reactions are executed in an event-driven manner. Spatiocyte, implemented as plugin algorithm modules to the E-Cell System (Arjunan and Tomita 2009; Arjunan 2009b), is a freely distributed open-source software. A guide to modeling with Spatiocyte is also available (Arjunan 2009a).
Verifying diffusion-influenced reactions
To verify the diffusion-influenced reactions of our method we employ a similar approach as in (Ridgway et al. 2008) by comparing the simulation results with the predicted survival probability of reacting species in irreversible (S irr) and reversible (S rev) reactions. Here, we describe the analytical and numerical methods to obtain S irr and S rev since they have not been previously described for surface reactions. Where possible we adopt the notations in (Popov and Agmon 2001).
Irreversible reactions
According to the Smoluchowski theory (von Smoluchowski 1917; Szabo 1989; Popov and Agmon 2001), in the irreversible reaction \(A+B\,\xrightarrow{k_{AB}}\,C\) with dilute A and B molecules, the fraction of A still surviving at time t is
where c 0 is the initial concentration of B and k irr(t) is the time-dependent rate coefficient. For volume reactions,
Here the function \(\Upphi(x)=\exp(x^2)\operatorname{erfc}(x),\) k D = 4π(D A + D B )R is the diffusion-limited rate constant, \(k_D'=k_{AB}k_D/(k_{AB}+k_D)\) is the asymptotic reaction rate and
For reactions taking place in the HCP lattice, the contact distance R = 2r v . In case of surface reactions, the time-dependent rate coefficient is only known in the Laplace space,
where
is the Green’s function for free 2D diffusion (Popov and Agmon 2002), obeying a reflective boundary condition at R; K n (z) is the modified Bessel function of the second kind for the integer order n; and τ D = R 2/(D A + D B ) is the diffusion-limited time constant. The transform (9) is numerically inverted to obtain k irr(t) and subsequently, the survival probability S irr(t;c 0).
Reversible reactions
For the reversible reaction \(A+B \underset{{k_{C} }}{\overset{{k_{{AB}} }}{\rightleftharpoons}} C\) with a single target A and many B molecules, the best known method to estimate the survival probability S rev(t) of A is the first variant of the multiparticle kernel theories (MPK1) (Sung and Lee 1999; Popov and Agmon 2001). The normalized deviation of S rev(t) from the predicted equilibrium value \(S_{\rm rev}(\infty) = k_D/\lambda,\) with λ = k C + c 0 k AB , is obtained by numerically calculating the inverse Laplace transform
Here, \(\tilde{F}_{\rm MPK1}(s)\) is the diffusion factor function of the reversible reaction according to MPK1,
where the modified concentration, \(c_0' =\lambda/k_{AB},\,\tilde{F}_{\rm gem}(s)=1+k_{AB}\tilde{G}(s)\) is the diffusion factor function for a geminate pair, while in the irreversible case, the function is given as
The reflective Green’s function in 2D is given in (10) whereas in 3D it is
\(\tilde{S}_{\rm irr}(s;c_0)\) in (13) is obtained by numerically computing the Laplace transform
Simulating fluorescence microscopy images
Spatial localization patterns of molecules both in vitro and in vivo are widely determined using fluorescence labeling experiments (Resch-Genger et al. 2008; Fernández-Suárez and Ting 2008; Ji et al. 2008; Huang et al. 2009). Co-localization patterns of multiple species of molecules can also be determined using multiple fluorescent labels. The merged localization images show the blending of colors when the positions of the separately labeled molecules overlap. However, the optical microscopy images suffer from low resolution because of the diffracted light and the long exposure time required by the camera in microphotography processes. The exposure time of several hundred miliseconds to several seconds makes it impossible to capture the position of diffusing molecules at single molecule resolution. As a result, it is difficult to directly compare the position of molecules obtained from spatial simulations with the microscopy images.
Here, we develop a visualization method that simulates microscopy images by displaying the trajectory of simulated molecules averaged according to the exposure time. We use the OpenGL programming interface for 3D computer graphics to develop the method. We adopt the RGBA mode of OpenGL, where the colors are represented by red, green, blue and alpha color components. The alpha color component allows a color to be blended with the existing color at the intended position. An alpha value of 0 implies full transparency, while an alpha value of 1 implies full opacity. The background color of the simulation space is set to black to represent the unlit space of the microscopy image. We record the voxel position of each molecule, regardless of its diffusion coefficient, at each minimum diffusion step interval τ m = min{τ i }. For each voxel, the number of times a molecule of a fluorescent-labeled species f has occupied it in the exposure time τ e can then be counted, and is given as N f . To display the trajectory of the labeled species f within the exposure time, the alpha value of each voxel is calculated as
while its RGB values are set to the species fluorescent color. The above step is repeated for the remaining fluorescent labeled species. When molecules of separately labeled species occupy the same voxel within the exposure time, or overlap during visualization, their colors are blended according to their opacity levels. In addition, the intensity of voxel colors is directly proportional to the density of molecules at the position during the exposure time, as it is the case in microscopy images.
Modeling the E-ring
To formulate a model of the MinDE system that can reproduce the E-ring, we looked at the properties of MinE. The domains at the two terminals of MinE appear to have separate roles in the polar zone oscillations although they have been found to interact with each other (Ramos et al. 2006). Firstly, in the absence of the MinE C-terminal domain, E-ring fails to appear (Raskin and de Boer 1997). The C-terminal also contains the homodimerization domain of MinE (King et al. 1999). Hence, effective homodimerizations of MinE may play a significant role in E-ring, especially since it is made up of dense MinE. Secondly, when E-rings fail to appear, MinD polar zones extend beyond the midcell and take much longer to cycle between the poles (Rowland et al. 2000), resulting in the minicelling phenotype. This implies that the MinD ATPase activity is substantially reduced (Huang et al. 2003) in the absence of E-ring as a consequence of the disrupted MinE C-terminal domain. Strikingly, the MinD interaction domain resides near the MinE N-terminal (Ma et al. 2003), which was present. Therefore, because the polar zone still appeared, the ATPase activation function in the MinE N-terminal domain is likely at a very low basal level when the C-terminal domain is disrupted. Thirdly, the truncated N-terminal domain, which forms a nascent helix (King et al. 1999), can autonomously bind to the membrane in the absence of MinD (Ma et al. 2003), and and in the full length MinE, the binding domain is thought to be exposed upon membrane recruitment by MinD (Ma et al. 2003). This further supports our theory that MinE constituting the E-ring remained transiently attached to the membrane independently after recruited by MinD.
We make two assumptions to account for the above MinE properties in our model. First, since MinE cannot independently associate to the membrane, the putative membrane binding domain near the MinE N-terminal is only exposed upon binding with MinD on the membrane as suggested by Ma et al. (2003). Second, we predict that MinE can only begin to transiently attach to the membrane independently of MinD when it is in the dimer form with the membrane binding domains at both N-terminals exposed. This is analogous to MinD, which must be in the dimer form for stable binding to the membrane (Zhou et al. 2005).
The series of reactions in the proposed model are as follows: (i) cytosolic MinDATP independently binds to the membrane, but with a very low affinity,
where the superscript m denotes that the molecule is membrane bound; (ii) MinDm cooperatively recruits cytosolic MinDATP,
(iii) MinDm also recruits cytosolic MinE dimer (represented as MinEE) to the membrane,
(iv) in the complex MinDEEm, MinE stimulates MinD ATPase function, triggering the release of MinD to the cytoplasm whereas MinEE remains transiently bound to the membrane via its exposed N-terminal membrane binding domains (Ma et al. 2003)
(v) the released MinDADP is phosphorylated in the cytoplasm,
(vi) the unbound MinE subunit in the MinDEEm complex can also associate with other MinD on the membrane,
(vii) the MinDEEDm complex releases MinDADP to the cytoplasm upon activation,
and (viii) because of the transient nature of MinEEm membrane association, it also dissociates to the cytoplasm
Note that the first five reactions, (17)–(21), are similar to the well known MinDE model by Huang et al. (2003). However, in reaction (20) of our scheme, the MinE dimer is transiently attached to the membrane instead of immediately becoming cytosolic after activating and releasing MinD. Table 2 lists the simulation parameters and their sources. We used the adapted NR method to execute reactions (20), (21), (23) and (24) since they are diffusion-independent. The remaining reactions were peformed using the proposed method for diffusion-influenced reactions.
Results
Diffusion in volume and surface compartments is verified
Figure 2 shows that the mean squared displacements of freely diffusing molecules in volume and surface compartments using Spatiocyte are indistinguishable from expected analytic values. The diffusion of the molecules is also correct when the walk probability P i /ρ i is less than unity in a diffusion process interval \(\tau_i'\). Based on the results in Fig. 2, the multi-timescale algorithm is also verified because the diffusion of all four species, in different compartment types and with unique combinations of diffusion process intervals and walk probabilities, were simultaneously and correctly simulated.
Verification of diffusion in volume and surface compartments. Solid lines show the mean squared displacement of molecules diffused according to (5) using the indicated parameters and averaged over 10000 runs with independent seeds. The lines almost perfectly overlap the predicted volume (dashed lines) and surface (dotted lines) diffusion results from analytic solutions. At the beginning of each run, molecules are placed randomly in their respective compartment. A cubic volume with periodic boundaries constitutes the volume compartment, while its six faces, covered with lipid molecules and imposed with periodic boundary condition at the edges, form the surface compartment. When a molecule walks over a periodic boundary, its absolute coordinate is updated to emulate borderless volume and surface compartments. All four species, with unique combinations of diffusion process interval \(\tau_i'\) and walk probability P i /ρ i , were simultaneously simulated to also verify the correctness of the multi-timescale algorithm. In both compartments, the excluded volume effect by the molecules is negligible because they occupy less than 0.01% of the simulated volume and surface area
Reactions in volume and surface compartments are verified
For the irreversible reaction \(A+B\,\xrightarrow{k_{AB}}\,C\) , it can be seen in Fig. 3a and b that the survival probabilities of A from simulations are in excellent agreement with the predicted theoretical curves in both type of compartments. The values of D A , D B and p j are chosen such that Spatiocyte is evaluated for reactions that are minimally to maximally diffusion-limited. In each compartment, the multi-timescale algorithm is also verified since the eight reactions were simultaneously performed correctly.
Verification of irreversible and reversible reactions in volume and surface compartments. a and b The survival probability of A in the second-order irreversible reaction \(A+B\,\xrightarrow{k_{AB}}\,C\), taking place in volume (a) and surface (b) compartments. Dashed curves are the predicted survival probability according to (6), while the solid colored curves are the simulation results averaged over 1000 independent iterations. In each compartment, eight reactions were simultaneously simulated with various combinations of diffusion coefficients, D A and D B , and the reactive collision probability p j , as indicated in the inset legend table. Each reaction involves 100 A and 100 B molecules, with the voxel radius r v = 0.5. For the volume and surface compartments, V = 1 × 106 and S = 3.5 × 105, respectively. c and d The normalized deviation of the survival probablity S rev(t) of A from the predicted equilibrium value S rev(∞) in the second-order reversible reaction \(A+B \underset{{k_{C} }}{\overset{{k_{{AB}} }}{\rightleftharpoons}}C\), taking place in volume (c) and surface (d) compartments. Black dashed and dotted curves are the predicted values calculated according to MPK1 theory (Sung and Lee 1999; Popov and Agmon 2001) in Eq. 11, dotted colored curves are theoretical predictions computed according to mass action, while the solid colored lines are the simulation results. The reaction and simulation parameters are indicated in the respective panels. In all reactions, the impact of molecular crowding is negligible because the molecules occupy less than 0.01% of the simulated volume and 0.8% of the surface area
Compared to irreversible reactions, reversible reactions present a harder test to spatial modeling methods because newly dissociated molecules that are close to each other have a higher tendency to recombine. We performed the reversible reaction \(A+B \underset{{k_{C} }}{\overset{{k_{{AB}} }}{\rightleftharpoons}}C\) with both weak (k C = 5) and strong (k C = 500) dissociation constants to evaluate the method. Since the dissociation reactions are diffusion-independent, they were executed using the adapted NR method. Figure 3c and d show the simulation results when the reactions take place in the volume and surface compartments, respectively. Taking into account the noise limits, the results are in good agreement with the best known approximation theory, MPK1 in both compartments, thus verifying the diffusion-independent and diffusion-influenced reactions of Spatiocyte. In addition, as previously reported in (Ridgway et al. 2008), mass action fails to correctly reproduce the predicted survival probability of A for volume reversible reactions (Fig. 3c) because it assumes A, B and C to be HD species. Figure 3d shows that mass action is also unable to reproduce the expected curves for surface reversible reactions.
Spatiocyte reproduces implications of molecular crowding
We evaluated Spatiocyte to reproduce the effects of volume exclusion by first examining the diffusion behavior of molecules in a crowded volume compartment. We mimicked the crowded Escherichia coli cytoplasm by occupying 34% of the 1 μm3 compartment volume (Zimmerman and Trach 1991) with inert and immobile crowder molecules. Figure 4a shows the time-dependent diffusion coefficients of six species that were diffused in the compartment. Of the six species, one is MinD with the diffusion coefficient 61 μm2 s−1, obtained by fluorescence correlation spectroscopy (FCS) in a dilute solution (Loose et al. 2008). The results indicate that the species undergo anomalous diffusion (i.e., sub-linear scaling of the mean-squared displacement over time) when the volume compartment is crowded. Corroborating the results of previous off-lattice simulations (Ridgway et al. 2008), the time-dependent diffusion coefficients reach equilibrium by 10−4 s, at which time they have decreased by about a factor of two compared to dilute levels.
Implications of molecular crowding in diffusion and reaction processes. a–c The time-dependent diffusion coefficients at 34% occupied volume (a), 34% occupied surface area (b) and 23% occupied surface area (c). The diffusion coefficients are calculated from the equation \(D(t)=<r^{2}>/(2lt)\), where \(<r^{2}>\) is the mean squared displacement and l is the spatial dimensionality. Line labels show the diffusion coefficients of six simulated species at dilute levels. d The survival probability of A in the volume irreversible reaction \(A+B\,\xrightarrow{k_{AB}}\,C\) at various occupied volume fractions (rounded to integer percentages). All simulation parameters are as indicated in the respective panels
Using the same six species, we investigated the diffusion behavior in two different cases of crowded surface compartment. In the first case (Fig. 4b), we occupied 34% of the surface area with crowders to allow direct comparison to the previously observed behavior in volume compartment. In the second case (Fig. 4c), 23% of the surface area was occupied by the crowders to reflect the recently determined in vivo value (Dupuy and Engelman 2008). The results of the first case show that the anomalous diffusion of the surface molecules is more pronounced than the observed behavior in the volume compartment. The elevated impact of crowding in the surface compartment is because of the increased obstructions to diffusion, resulting from the smaller dimensionality of the surface space (Zhou 2009). Interestingly in the second case, although the area occupancy is 23%, the surface diffusion coefficient at equilibrium (t = 10−4 s) is almost equivalent to that of the volume at 34% occupancy.
We evaluated the effects of molecular crowding in volume irreversible reactions. Figure 4d shows that the apparent survival probability of the reacting molecules increases in line with the volume occupancy. There are more surviving reactants at higher occupancy because their time-dependent diffusion coefficient is reduced by the increased hindrances from crowders. Together, these results confirm that our method can account for some of the important implications of molecular crowding in volume and surface compartments.
Spatiocyte visualization method simulates microscopy images
Figure 5 compares the localization patterns of simulated molecules at single molecule resolution to the corresponding simulated fluorescence microscopy image using the Spatiocyte visualization method. The radius of the molecules, occupying an Escherichia coli membrane, is 10 nm while the exposure time of the simulated image is 500 ms. Since the molecules are very small, it is difficult to identify the patterns at single molecule resolution. Conversely, the patterns are clearly defined in the simulated fluorescence image because the overall trajectory of molecules during the exposure time is shown. Co-localizing molecules can also be identified by their blended colors.
Comparison of single molecule visualization to simulated microscopy image. a Single molecule visualization (Top) of MinDEm (yellow) and MinEm (cyan) on Escherichia coli membrane compared to the corresponding simulated fluorescence microscopy image (Bottom). The exposure time of the fluorescence image is 500 ms. The proteins were simulated according to reactions (17)–(24). The diffusion coefficients and model parameters are listed in Table 2. b The magnified area of the respective boxes shown in (a)
Comparisons of actual microscopy images to the corresponding simulated images are shown in Fig. 6c. The left column of the panel illustrates the in vivo localization patterns of MinD and MinE from the doubly labeled experiments by Shih et al. (2002). Using an exposure time of 500 ms, the Spatiocyte visualization method closely reproduces the microscopy images in both individual and overlay panels. However, the simulated patterns are not as smooth as in the microscopy images possibly because the diffraction of light was not considered. It is also evident that the visualizations of single molecules do not clearly show the localization patterns as observed in the microscopy images.
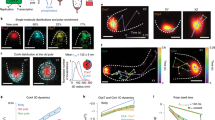
Comparison of MinDE polar zone and E-ring oscillations in wild type and mutant cells. a Oscillations of MinDm (orange), MinDEm (purple) and MinEm (cyan) with transient membrane attachment of MinEm according to the proposed reactions (17)–(24) (see Table 2 for model parameters). b Same color scheme as in (a) but employing the adapted Huang et al. (2003) model (reactions (17)–(21)). In the Huang et al. model, reaction (20) is altered such that MinE is directly released to the cytoplasm together with MinD upon ATP hydrolysis instead of transiently attaching to the membrane. The reaction rates were fitted to reproduce the pole-to-pole oscillations (see text). MinE is represented as monomers in the reactions for simplicity. c Comparison of wild type in vivo localization patterns of MinD and MinE (Left Column) to simulation (Right Column) according to our proposed model in (a). In the simulation panels, the corresponding simulated microscopy image (Upper) and single molecule visualization (Lower) are shown. In the bottom simulation panel, the molecule color scheme is as in (a). d Comparison of in vivo dynamic localization patterns of Gfp-MinD (Top Row) to simulation (Bottom Row) in a mutant strain expressing MinE1-53, where the C-terminal region of MinE comprising amino acids 54–88 was removed. For the simulation, the color scheme and the proposed model of MinE transient membrane association in (a) were used but the MinD ATPase activation rates k 4 and k 7 were reduced to 0.06 s−1. The length of the simulated cell was increased to 8 μm, estimated from the mutant. (c) (Left Column) is adapted by permission from Macmillan Publishers Ltd: The EMBO Journal (Shih et al. 2002), © 2002. (d) (Top Row) is reproduced with permission from (Rowland et al. 2000) © 2000, American Society for Microbiology
A wide range of model parameters reproduce MinDE oscillation dynamics
The stochastic simulation results of our wild type MinDE model are presented in Fig. 6a with the corresponding movie in Video S1. The dynamic localization patterns of the oscillation are in agreement with those observed in experiments (Raskin and de Boer 1999; Shih et al. 2002). Morever, dense MinEm can be seen clearly localizing annularly to form the E-ring at the edge of the polar zones (see also Fig. 6c). The average oscillation period for 20 cycles is 48 ± 6 s, which is consistent with the experimentally measured average value by Shih et al. (2002). The oscillation period can also be increased to be within the range of 50 s to 120 s as experimentally determined by Meacci and Kruse (2005) by reducing the MinE transient membrane dissociation rate, k 8. For the model, 1 min of simulated time takes approximately 4 min of runtime on an Intel Core 2 Extreme 3.2 GHz system with 8 GB of RAM.
The parameters that we used for the model were obtained from experimental data if available, otherwise they were either fitted or approximated (see Table 2). Significantly, compared to the MinD nucleotide exchange rate used in previous models (Huang et al. 2003; Kerr et al. 2006; Fange and Elf 2006), ours was increased by several folds to 5 s−1 to meet the lower bound determined by Meacci et al. (2006). Furthermore, increasing or reducing the fitted reaction rates simultaneously by twofold did not substantially affect the oscillation patterns but changed the average oscillation periods to 24 ± 4 and 116 ± 23 s, respectively. It is also possible to generate the oscillations and E-rings when the changes are larger than twofold but each fitted reaction rate needed to be readjusted. The initial positions of the molecules in the cytoplasm or on the membrane also did not have an effect in establishing and sustaining the oscillations. In addition, reducing the cell length from 4.5 to 3.5 μm or increasing the length to 5.5 μm still maintained the dynamic localization patterns of MinD and MinE with the average oscillation periods of 47 ± 6 and 55 ± 8 s, respectively. Taken together, a wide range of parameter values can be used to reproduce the E-ring and MinDE oscillation dynamics with our model.
Transient membrane attachment of MinE leads to the formation of E-ring
Figure 6c shows that the wild type localization patterns of MinE and MinD from our simulations are consistent with the in vivo results obtained by Shih et al. (2002). The simulated E-ring, nonetheless, is more dispersed than its in vivo counterpart possibly because we did not consider MinE oligomerization (Pichoff et al. 1995; Zhang et al. 1998) and/or MinD polymerization (Shih et al. 2003), both of which can substantially reduce the diffusion coefficient and sharply increase the subunit density at the localization site. In addition, a significant portion of the E-ring is composed of MinE that is transiently attached to the membrane independent of MinD (shown in cyan in Fig. 6a and c). We conclude that our wild type model can reproduce the E-ring similar to the observations in vivo.
To confirm that our model can also reproduce the mutant phenotypes of Escherichia coli, we compared simulation results with in vivo dynamic localization patterns of MinD and MinE when the C-terminal domain of MinE is disrupted. As evident in Fig. 6d, our model accurately reproduced the disappearance of E-ring and the diffusely extended MinD polar zones observed by Rowland et al. (2000) when we reduced the MinD ATPase activation rates k 4 and k 7 to 0.06 s−1. Additionally, the oscillation also has a very long period of ≈10 min, which corresponds to what has been observed experimentally (Rowland et al. 2000). Our results agree with previous suggestions by Huang et al. (2003) and Derr et al. (2009) that the reduction of the ATP hydrolysis rate can increase the period and reduce the intensity of E-rings. In our model, however, the E-ring is no longer visible because MinEm is dissociated much faster (k 8) than its effective recruitment rates (arising from k 4 and k 7). MinE was represented as homodimers in the model although it likely existed as monomers in vivo since the C-terminal domain, required for homodimerization, was removed (Rowland et al. 2000). As a result, we readjusted the model to represent MinE as monomers, which still reproduced the mutant phenotypes when the hydrolysis rate was decreased (data not shown).
In both experiment (Shih et al. 2002) and in our simulations, the growing polar zone appears to be blocked at midcell by the E-ring before shrinking and subsequently growing again at the opposite pole. The polar zone grows because of independent nucleation and cooperative recruitment of MinDATP to the membrane. The shrinkage is the result of MinDm stimulation by the MinE population from the cytoplasm that is recruited to the membrane. Adding another reaction to the wild type model that allows MinEm to bind and stimulate MinDm on the membrane did not appreciably alter the dynamic localization patterns (data not shown). Therefore in vivo, the transiently attached MinEEm could also be associating with MinDm directly on the membrane, bypassing the cytosolic phase.
To see if the E-ring is still generated as observed in vivo when MinE is immediately released to the cytoplasm instead of being transiently attached to the membrane upon ATP hydrolysis, we employed the adapted Huang et al. (2003) model, comprising only reactions (17)–(21). MinE was represented as monomers instead of dimers in the reactions to simplify the model and as a result, the remaining three reactions (22)–(24) were removed. In the adapted reaction (20), we replaced the product MinEm with cytosolic MinE to exclude transient membrane attachments of MinE. The reaction rates k 1 (0.035 μm s−1), k 2 (0.04 μm3 s−1), k 3 (0.5 μm3 s−1), k 4 (0.15 s−1) and k 5 (5 s−1) were fitted to reproduce the pole-to-pole oscillations. The resulting dynamic localization patterns of MinDE are shown in Fig. 6b. The results indicate that MinDEm localized annularly at the edge of polar zones that cycled with an average period of 71 ± 8 s. Because the simulated MinE was always found dimerized with MinD on the membrane, the in vivo localization pattern of E-ring, which is not co-localized with MinD as shown in Fig. 6c (Shih et al. 2002), was not reproduced. Hence, we conclude that MinE must be able to transiently attach to the membrane independently once recruited by MinD to reproduce the E-ring.
Width of E-ring is proportional to MinD ATPase activation rate
The basic factors that establish and sustain in vivo MinDE oscillations according to the Huang et al. model have previously been described (Huang et al. 2003; Arjunan and Tomita 2009). Here we examine the oscillation characteristics in terms of the adapted reaction (20) and the three new reactions (22)–(24) that we added to the model to generate the E-ring.
As shown in the previous section, the E-ring is formed as result of the transient membrane association of MinEEm after MinD is dissociated to the cytoplasm in reactions (20) and (23). Increasing the rates of ATP hydrolysis, k 4 and k 7, widened the E-ring and decreased the oscillation period. This is expected because more transiently membrane-associated MinEEm are available to form the E-ring while MinD in the polar zones is rapidly removed. Conversely, the oscillations become slower with thinner and fainter E-rings while the polar zones elongated when the rates were reduced. The fainter E-rings are consistent with the in vivo localization patterns resulting from a point mutation to the MinE C-terminal domain (MinED45A) conducted by Shih et al. (2002). Our results thus imply that the mutation at the C-terminal domain may have impaired the MinD activation function of MinE.
Increasing the rate of reaction (22) narrowed the E-ring by reducing the subsequent production of transiently bound MinEEm dimers but decreased the period. The oscillation is rapid because more MinDEEm associates with MinDm that are eventually activated. In reaction (24), increasing the rate MinEEm is dissociated to the cytoplasm also had a similar effect in that it narrowed the E-ring and made the oscillation faster. The burst in the cytosolic MinE population that are capable of rapidly rebinding with MinDm contributes to the shortening of the period. Reciprocally, E-rings widen with slower oscillations when the dissociation rate is reduced. However, if the rate is too small the E-rings become dispersed as a result of MinEEm lateral diffusion.
Overall, the population of MinDEEm, MinDEEDm and MinEEm are inversely proportional to the rates k 4, k 7 and k 8 respectively. The ratio of the three rates with respect to each other also generally determines the population ratio of the three different MinE species on the membrane. For example, higher k 4 and k 7 than k 8 will generate less MinDEEm and MinDEEDm compared to MinEEm.
Discussion
A new spatio-temporal modeling method
We developed and verified a new lattice-based method, Spatiocyte that can model surface and volume intercompartmental RD processes, while also being able to account for the implications of stochasticity and molecular crowding arising from the physical dimensions of molecules. To our knowledge, there is currently no single method that can do this (see Table 1). We have also adopted several optimization strategies to reduce the high computational cost associated with spatial modeling methods. Firstly, unlike other methods, Spatiocyte can simultaneously simulate highly concentrated or HD species at compartmental scale and low copy or heterogeneously distributed species at molecular scale. This allows the method to skip diffusing each molecule belonging to the highly concentrated species, which would incur very high computational cost. Molecules within the two distinct spatial scales can react as usual with each other. Secondly, the reactions involving HD species are event-driven so that computations are performed only when necessary, thus allowing bigger jumps in the simulation time. Thirdly, to support fast RD simulation in crowded compartments, the simulation space is discretized into hexagonal close-packed lattice that enables rapid resolving of molecular collisions. The overall reduced computational cost is evident as it only takes approximately 4 min to simulate 1 min of the MinDE system using Spatiocyte on a desktop computer.
The obvious limitation of Spatiocyte is that the non-HD molecules have to be almost similar in size to occupy the voxels. This presents a problem when modeling molecules with disparate densities in a crowded compartment since the size of the molecules directly influences RD. Nonetheless, in most spatial modeling applications, the molecules are assumed to be uniform sized. In addition, the method can also be extended to model molecules of different sizes by occupying multiple voxels with a single molecule according to its size. In this case, the reactive collision probability will need to be readjusted to account for the collisions that take place at the multiple voxels occupied by the molecule.
We also developed a visualization method that uses the temporal positional data of simulated molecules to simulate optical microscopy images of fluorescent labeled proteins. It does this by showing the trajectory of simulated molecules averaged according to the camera exposure time used in the microphotography process. The method enabled us to directly match our simulation results with the localization patterns of MinD and MinE in Escherichia coli and make objective evaluations. Schaub et al. (2007) have also developed a method to simulate the microscopy images but it is intended for almost immobile, non-diffusing structures, such as actin filaments, and cannot be directly applied for diffusing molecules as required by our method.
In our results, the 34% crowded volume compartment caused the diffusion coefficient of MinD to be reduced by a factor of two to approximately 37 μm2 s−1 from its dillute level of 61 μm2 s−1 (Loose et al. 2008). However, the measured diffusion coefficient of MinD in the 34% crowded Escherichia coli cytoplasm is only 16 μm2 s−1 (Meacci et al. 2006). The additional twofold reduction can be attributed to the several factors listed by Ridgway et al. (2008), which include the intracellular viscosity and the nonspecific binding to other molecules in the cytoplasm. This suggests that adding crowder molecules alone may not be sufficient to reproduce the cytoplasmic environment in a simulated model.
Our results also support the suggestion by Loose et al. (2008) that molecular crowding on the cell membrane decreases the lateral diffusion coefficients of MinD and MinE compared to that of in vitro. Additional reduction in the diffusion coefficients could also be attributed to the protein oligomerization and polymerization-depolymerization kinetics on the membrane. Polymers on the membrane may also present as bigger hindrances to the mobility of the proteins.
A new MinDE model that reproduces the E-ring
In the second part of this study, we modeled the MinDE dynamics using Spatiocyte to elucidate the mechanism of E-ring formation, which remains ambiguous. Based on the high density of MinE in the E-ring that is not co-localized with MinD at least half of that density (assuming each MinD subunit recruited a MinE dimer at the E-ring) (Shih et al. 2002), and the ability of the truncated N-terminal domain of MinE to bind autonomously to the membrane Ma et al. (2003), we predicted that the MinE constituting the E-ring remains transiently bound to the membrane autonomously after being recruited by MinD. To evaluate our prediction, we incorporated transient membrane attachments of MinE (MinEm) in our MinDE model.
We reduced the complexity and parameters of the model by incorporating a small number of reactions that are necessary to reproduce the MinDE dynamics observed in vivo. Consequently, the reaction and diffusion parameters are effective parameters that might represent multiple processes such as oligomerization and polymerization, which are not explicitly represented. We have also made two assumptions in the model. First, as suggested by Ma et al. (2003), we assume that the putative membrane binding domain near the MinE N-terminal is only exposed upon binding with MinD on the membrane. Second, we speculate that MinE must be in a dimer form with both N-terminal membrane binding domains exposed by MinD before it can transiently attach to the membrane independently of MinD.
With the simplification and assumptions, our model reproduced the MinDE oscillation dynamics and E-ring that are consistent with the observations in vivo. Using the Spatiocyte visualization method, we can directly compare the in vivo co-localization patterns of doubly labeled MinD and MinE (Shih et al. 2002) with simulation. Significantly, our results for the first time show that the transient membrane attachment of MinE is necessary to reproduce the E-ring as observed in vivo. As we predicted, a significant portion of the simulated E-ring consisted of transiently membrane attached MinEm. The results also indicate that the width of E-ring is proportional to the MinD ATPase activation rate and inversely proportional to the membrane detachment rate of MinEm. Higher binding rates of MinDEEm to MinDm also narrowed the E-ring. When the C-terminal domain of MinE was removed, the model recovered the in vivo mutant phenotypes, including the longer polar zones, the substantial delay in the oscillation period and the loss of E-ring. Our simulation results attribute the mutant phenotypes to the reduction of MinD ATPase activation by MinE.
We suggest a possible scenario for the reduced ATPase activation rate. In the mutant, MinE is mostly found in the form of monomers because the domain responsible for dimerization near the C-terminal region has been removed. Wild type MinE in the dimer form could activate two MinD monomers concurrently and thus, it is more effective in activating multiple MinD promoters at a localized site of an oligomer or a polymer on the membrane. For example, at the leading edge of the polar zone where the E-ring forms, it is crucial for MinE to dissociate MinD faster than it is recruited to the membrane. Otherwise, it would not be possible to rapidly shrink the polar zone as seen in the wild type cells. Therefore, when the homodimerization of MinE1−53 is impaired, the effective ATPase activation rate could be significantly reduced, resulting in the mutant phenotypes.
Interestingly, MinE C-terminal point mutations of the residues Asp45 and Val49 to Ala in MinED45A/V49A also resulted in the minicelling phenotype (King et al. 2000; Shih et al. 2002) and the complete loss of E-rings although MinED45A/V49A homodimerizes in vitro (Shih et al. 2002). The mutated residues form a highly conserved tetrad at the center of the MinE dimer interface (see Fig. 7a and King et al. 2000). Even though MinED45A/V49A might be in the dimer form in the cytoplasm and in vitro conditions that are free of membrane and MinD (Shih et al. 2002), we predict that the dimer could be destabilized and dissociated on the cell membrane by its N-terminal interactions with dimeric or polymeric MinD because of the weakened mutated dimeric bonds. As shown in the secondary structure of the dimer in Fig. 7b, the mutated tetrad is located at the center of solvent exposed face of the alpha helices and might be essential for the dimer stability when it associates with MinD. The proximity of the tetrad to the N-terminal domains indicates that it could be subjected to immediate stress as MinD is associated at both ends, which could be further amplified when MinD forms higher-order complexes. Figure 7c illustrates an example of MinDE interaction model with the destabilization of MinED45A/V49A dimer by MinD. Additionally, a single point mutation of Asp45 to Ala in MinED45A was capable of generating narrower E-rings, which we could attribute to a less destabilized MinED45A dimer interface. Parallel to the narrowing of the E-ring width when we decreased the activation rates k 4 and k 7 in our simulation, some MinED45A that are able to stay dimerized on the membrane could strongly activate MinD and form the narrower E-rings.
The \(MinE^{D45A/A49A}\) mutant dimer and molecular models of MinE interactions. a Space-filling representation of C-terminal MinE31-88 homodimeric structure showing the positions of Asp45 (magenta) and Val49 (light green) residues that form a highly conserved tetrad at the center. b The corresponding secondary structure of MinE31-88 homodimer as shown in (a). The two monomers are distinguished by the colors brown and dark green. The magenta and light green patches on the helices show the positions of Asp45 and Val49 respectively. c Molecular models of wild type MinE and MinED45A/A49A interactions in the MinDE system. The models are represented by MinD (lime green box) that can be a subunit of an oligomer/polymer (gray box), MinE N-terminal (EN) and C-terminal (EC) domains, inner membrane (light blue horizontal block), cytoplasm (white space), membrane binding domain (thick dark blue line), dimerization bond (thick black line), Asp45 (magenta box) and Val49 (light green box) dimerization interfaces, dissociation reaction (black arrow) and transition of reactions (red arrow). (Wild Type MinE)(i) Cytosolic MinE, mostly existing as dimers, is recruited to the membrane by MinD oligomers. The membrane binding domain at the N-terminals of the dimer are exposed by MinD upon recruitment. (ii) The MinE dimer effectively activates MinD because both of its N-terminal domains are associated with MinD at a localized site of the MinD oligomer or polymer. Once MinD is released to the cytoplasm, the dimer remains transiently bound to the membrane via the membrane binding region at both of its N-terminals. The transiently bound dimer could either dissociate to the cyoplasm (iii) or associate with new MinD protomers (iv). (v) MinE dimer also activates the ATPase function of a single associated MinD but at a reduced effective rate. (MinED45A/A49A Mutant)(i) In the cyoplasm, MinED45A/A49A may exist as a homodimer or a monomer. The homodimer (or two monomers) is recruited to the membrane by MinD oligomers. The membrane-binding domain at the N-terminal of MinED45A/A49A is also exposed by MinD. (ii) The recruited MinED45A/A49A dimer is destabilized on the membrane by MinD and dissociates. (iii) The effective MinD activation rate by the dissociated MinED45A/A49A monomers is reduced since they are not in a dimer form at a localized site. Upon activation, both MinD and MinED45A/A49A monomers are released into the cytoplasm. MinED45A/A49A is not transiently bound to the membrane because it must be in the dimer form with the membrane binding domain of both monomers exposed to support transient binding
Since a wide range of our model parameters was able to reproduce the dynamic localization patterns of MinDE, the model could also approximate more elaborate models comprising detailed reactions such as MinDE oligomerization. In Fig. 7c we illustrate two such detailed models of wild type MinE and MinED45A/V49A mutant interactions in the MinDE system. Recently, Derr et al. (2009) proposed a partial non-oscillating polymerization model of the MinDE system that do not explicitly include cooperative activation of MinD to reproduce E-rings. Because in the model MinE is always found dimerized with MinD on the membrane, it cannot recover E-rings that are much denser than MinD in the polar zone. It will be interesting to see how polymerization of MinD affects the formation of E-ring when transient membrane attachment of MinE is incorporated. Since stronger ATPase activation would be required to fragmentize and depolymerize annealing MinD polymers than freely diffusing MinD as the case in our model, we predict denser E-ring would form at the rim of the polar zones if we extend our model to include MinD polymerization.
By observing E-ring localization patterns in fluorescence microscopy images, it is difficult to determine what causes such dynamic annular structure formation. Based on the density of MinE in the E-ring that is much higher than MinD at the rim of polar zones, we can predict that it could be caused by transient membrane attachment of MinE. As we showed in this work, using Spatiocyte to create a model based on the known properties of MinE and the previously reported wild type and mutant localization patterns of the MinDE system, we can substantiate our prediction. Moreover, the model also provided further mechanistic insights about the system, such as the width of E-ring could be proportional to the rate of MinD activation.
References
Adams DW, Errington J (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7(9):642–653
Ander M, Beltrao P, Ventura BD, Ferkinghoff-Borg J, Foglierini M, Kaplan A, Lemerle C, Tomas-Oliveira I, Serrano L (2004) SmartCell, a framework to simulate cellular processes that combines stochastic approximation with diffusion and localisation: analysis of simple networks. Syst Biol 1(1):129–138
Andrews SS, Bray D (2004) Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys Biol 1(3–4):137–151
Arjunan SNV (2009a) A guide to modeling reaction-diffusion of molecules with the E-Cell System. In: Arjunan SNV, Dhar PK, Tomita M (eds) E-Cell System: basic concepts and applications. Landes Bioscience and Springer Science+Business Media (in press)
Arjunan SNV (2009b) Modeling three-dimensional spatial regulation of bacterial cell division. PhD Thesis, Keio University
Arjunan SNV, Tomita M (2009) Modeling reaction-diffusion of molecules on surface and in volume spaces with the E-Cell System. Int J Comput Sci Inf Secur 3(1):211–216
Balgi G, Leckb D, Nitsche J (1995) Transport effects on the kinetics of protein-surface binding. Biophys J 68(6):2251–2260
Bernhardt TG, de Boer PAJ (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18(5):555–564
Borowski P, Cytrynbaum EN (2009) Predictions from a stochastic polymer model for the MinDE protein dynamics in Escherichia coli. Phys Rev E 80(4):041916
Boulianne L, Assaad SA, Dumontier M, Gross W (2008) GridCell: a stochastic particle-based biological system simulator. BMC Syst Biol 2(1):66
Clarke DC, Liu X (2008) Decoding the quantitative nature of TGF-beta/Smad signaling. Trends Cell Biol 18(9):430–442
Collins FC, Kimball GE (1949) Diffusion-controlled reaction rates. J Colloid Sci 4(4):425–437
Cytrynbaum EN, Marshall BDL (2007) A multistranded polymer model explains MinDE dynamics in E. coli cell division. Biophys J 93(4):1134–1150
Derr J, Hopper JT, Sain A, Rutenberg AD (2009) Self-organization of the MinE protein ring in subcellular Min oscillations. Phys Rev E 80(1 Pt 1):011922
Dix JA, Verkman AS (2008) Crowding effects on diffusion in solutions and cells. Annu Rev Biophys 37:247–263
Drew DA, Osborn MJ, Rothfield LI (2005) A polymerization-depolymerization model that accurately generates the self-sustained oscillatory system involved in bacterial division site placement. Proc Natl Acad Sci USA 102(17):6114–6118
Dupuy AD, Engelman DM (2008) Protein area occupancy at the center of the red blood cell membrane. Proc Natl Acad Sci USA 105(8):2848–2852
Elf J, Ehrenberg M (2004) Spontaneous separation of bi-stable biochemical systems into spatial domains of opposite phases. Syst Biol 1(2):230–236
Ellis RJ (2001) Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci 26(10):597–604
Fange D, Elf J (2006) Noise-induced Min phenotypes in E. coli. PLoS Comput Biol 2(6):e80
Fernandez-Suarez M, Ting AY (2008) Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol 9(12):929–943
Gibson M, Bruck J (2000) Efficient exact stochastic simulation of chemical systems with many species and many channels. J Phys Chem A 104(9):1876–1889
Gillespie DT (1976) A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Comput Phys 22(4):403–434
Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81(25):2340–2361
Gomez-Uribe CA, Verghese GC (2007) Mass fluctuation kinetics: capturing stochastic effects in systems of chemical reactions through coupled mean-variance computations. J Chem Phys 126(2):024109
Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J (2009) Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol 7(6):e1000137
Hall D, Minton A(2003) Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim Biophys Acta 1649(2):127–139
Hansen CH, Endres RG, Wingreen NS (2008) Chemotaxis in Escherichia coli: a molecular model for robust precise adaptation. PLoS Comput Biol 4(1):e1
Hattne J, Fange D, Elf J (2005) Stochastic reaction-diffusion simulation with MesoRD. Bioinformatics 21(12):2923–2924
Howard M, Rutenberg AD (2003) Pattern formation inside bacteria: fluctuations due to the low copy number of proteins. Phys Rev Lett 90(12):128102
Howard M, Rutenberg AD, de Vet S (2001) Dynamic compartmentalization of bacteria: accurate division in E. coli. Phys Rev Lett 87(27 Pt 1):278102
Hu Z, Lutkenhaus J (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 34(1):82–90
Hu Z, Lutkenhaus J (2001) Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell 7(6):1337–1343
Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA 96(26):14819–14824
Hu Z, Saez C, Lutkenhaus J (2003) Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J Bacteriol 185(1):196–203
Huang B, Bates M, Zhuang X (2009) Super-resolution fluorescence microscopy. Annu Rev Biochem 78:993–1016
Huang KC, Meir Y, Wingreen NS (2003) Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones. Proc Natl Acad Sci USA 100(22):12724–12728
Ideker T, Lauffenburger D (2003) Building with a scaffold: emerging strategies for high- to low-level cellular modeling. Trends Biotechnol 21(6):255–262
Ji N, Shroff H, Zhong H, Betzig E (2008) Advances in the speed and resolution of light microscopy. Curr Opin Neurobiol 18(6):605–616
Kerr RA, Levine H, Sejnowski TJ, Rappel W (2006) Division accuracy in a stochastic model of Min oscillations in Escherichia coli. Proc Natl Acad Sci USA 103(2):347–352
King GF, Rowland SL, Pan B, Mackay JP, Mullen GP, Rothfield LI (1999) The dimerization and topological specificity functions of MinE reside in a structurally autonomous C-terminal domain. Mol Microbiol 31(4):1161–1169
King GF, Shih YL, Maciejewski MW, Bains NP, Pan B, Rowland SL, Mullen GP, Rothfield LI (2000) Structural basis for the topological specificity function of MinE. Nat Struct Biol 7(11):1013–1017
Klann MT, Lapin A, Reuss M (2009) Stochastic simulation of signal transduction: impact of the cellular architecture on diffusion. Biophys J 96(12):5122–5129
Kruse K (2002) A dynamic model for determining the middle of Escherichia coli. Biophys J 82(2):618–627
Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P (2008) Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320(5877):789–792
Lutkenhaus J (2007) Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76:539–562
Ma L, King G, Rothfield L (2003) Mapping the MinE site involved in interaction with the MinD division site selection protein of Escherichia coli. J Bacteriol 185(16):4948–4955
Meacci G, Kruse K (2005) Min-oscillations in Escherichia coli induced by interactions of membrane-bound proteins. Phys Biol 2(2):89–97
Meacci G, Ries J, Fischer-Friedrich E, Kahya N, Schwille P, Kruse K (2006) Mobility of Min- proteins in Escherichia coli measured by fluorescence correlation spectroscopy. Phys Biol 3(4):255–263
Meinhardt H, de Boer PA (2001) Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci USA 98(25):14202–14207
Minton A (2001) The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem 276(14):10577–10580
Morris DM, Jensen GJ (2008) Toward a biomechanical understanding of whole bacterial cells. Annu Rev Biochem 77:583–613
Neves SR, Iyengar R (2009) Models of spatially restricted biochemical reaction systems. J Biol Chem 284(9):5445–5449
Neves SR, Tsokas P, Sarkar A, Grace EA, Rangamani P, Taubenfeld SM, Alberini CM, Schaff JC, Blitzer RD, Moraru II, Iyengar R (2008) Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell 133(4):666–680
Pavin N, Paljetak HC, Krstict V (2006) Min-protein oscillations in Escherichia coli with spontaneous formation of two-stranded filaments in a three-dimensional stochastic reaction-diffusion model. Phys Rev E 73(2 Pt 1):021904
Pichoff S, Vollrath B, Touriol C, Bouche JP (1995) Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol Microbiol 18(2):321–329
Popov AV, Agmon N (2001) Three-dimensional simulations of reversible bimolecular reactions: The simple target problem. J Chem Phys 115(19):8921–8932
Popov AV, Agmon N (2002) Exact solution for the geminate ABCD reaction. J Chem Phys 117(12):5770–5779
Ramos D, Ducat T, Cheng J, Eng NF, Dillon JR, Goto NK (2006) Conformation of the cell division regulator MinE: evidence for interactions between the topological specificity and anti-MinCD domains. Biochemistry 45(14):4593–4601
Rangamani P, Iyengar R (2008) Modelling cellular signalling systems. Essays Biochem 45:83–94
Rao CV, Ordal GW (2009) The molecular basis of excitation and adaptation during chemotactic sensory transduction in bacteria. Contrib Microbiol 16:33–64
Raskin DM, de Boer PA (1997) The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91(5):685–694
Raskin DM, de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96(9):4971–4976
Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T (2008) Quantum dots versus organic dyes as fluorescent labels. Nat Methods 5(9):763–775
Ridgway D, Broderick G, Ellison MJ (2006) Accommodating space, time and randomness in network simulation. Curr Opin Biotechnol 17(5):493–498
Ridgway D, Broderick G, Lopez-Campistrous A, Ru’aini M, Winter P, Hamilton M, Boulanger P, Kovalenko A, Ellison MJ (2008) Coarse-grained molecular simulation of diffusion and reaction kinetics in a crowded virtual cytoplasm. Biophys J 94(10):3748–3759
Rodriguez JV, Kaandorp JA, Dobrzyki M, Blom JG (2006) Spatial stochastic modelling of the phosphoenolpyruvate-dependent phosphotransferase (PTS) pathway in Escherichia coli. Bioinformatics 22(15):1895–1901
Rowland SL, Fu X, Sayed MA, Zhang Y, Cook WR, Rothfield LI (2000) Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J Bacteriol 182(3):613–619
Sanford C, Yip MLK, White C, Parkinson J (2006) Cell++-simulating biochemical pathways. Bioinformatics 22(23):2918–2925
Schaff JC, Slepchenko BM, Choi Y, Wagner J, Resasco D, Loew LM (2001) Analysis of nonlinear dynamics on arbitrary geometries with the Virtual Cell. Chaos 11(1):115–131
Schaub S, Meister J, Verkhovsky AB (2007) Analysis of actin filament network organization in lamellipodia by comparing experimental and simulated images. J Cell Sci 120(Pt 8):1491–1500
Shih Y, Fu X, King GF, Le T, Rothfield L (2002) Division site placement in E. coli: mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J 21(13):3347–3357
Shih Y, Le T, Rothfield L (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA 100(13):7865–7870
Stefan MI, Edelstein SJ, Novere NL (2008) An allosteric model of calmodulin explains differential activation of PP2B and CaMKII. Proc Natl Acad Sci USA 105(31):10768–10773
Stiles JR, Bartol TM (2001) Monte Carlo methods for simulating realistic synaptic microphysiology using MCell. In: Schutter ED (ed) Computational neuroscience: realistic modeling for experimentalists, CRC Press, Boca Raton, pp 87–127
Sung J, Lee S (1999) Nonequilibrium distribution function formalism for diffusion-influenced bimolecular reactions: Beyond the superposition approximation. J Chem Phys 111(3):796–803
Szabo A (1989) Theory of diffusion-influenced fluorescence quenching. J Phys Chem 93(19):6929–6939
Szpiro GG (2003) Kepler’s conjecture: how some of the greatest minds in history helped solve one of the oldest math problems in the world, 1st edn. Hoboken, New Jersey
Takahashi K, Kaizu K, Hu B, Tomita M (2004) A multi-algorithm, multi-timescale method for cell simulation. Bioinformatics 20(4):538–546
Takahashi K, Arjunan SNV, Tomita M (2005) Space in systems biology of signaling pathways—towards intracellular molecular crowding in silico. FEBS Lett 579(8):1783–1788
Takahashi K, Tanase-Nicola S, ten Wolde PR (2009) Spatio-temporal correlations can drastically change the response of a MAPK pathway. Proc Natl Acad Sci USA (in press)
Tostevin F, Howard M (2006) A stochastic model of Min oscillations in Escherichia coli and Min protein segregation during cell division. Phys Biol 3(1):1–12
Treanor B, Batista FD (2007) Mechanistic insight into lymphocyte activation through quantitative imaging and theoretical modelling. Curr Opin Immunol 19(4):476–483
van Albada SB, ten Wolde PR (2009) Differential affinity and catalytic activity of CheZ in E. coli chemotaxis. PLoS Comput Biol 5(5):e1000378
van Zon JS, ten Wolde PR (2005) Green’s-function reaction dynamics: a particle based approach for simulating biochemical networks in time and space. J Chem Phys 123(23):234910
Vats P, Yu J, Rothfield L (2009) The dynamic nature of the bacterial cytoskeleton. Cell Mol Life Sci 66(20):3353–3362
von Smoluchowski M (1917) Versuch einer mathematischen Theorie der Koagulationskinetik kolloider Lsungen. Z Phys Chem, Stoechiom Verwandtschaftsl 92:129–168
Wils S, Schutter ED (2009) STEPS: modeling and simulating complex reaction-diffusion systems with Python. Front Neuroinformatics 3:15
Woldringh CL, Mulder E, Valkenburg JA, Wientjes FB, Zaritsky A, Nanninga N (1990) Role of the nucleoid in the toporegulation of division. Res Microbiol 141(1):39–49
Woldringh CL, Mulder E, Huls PG, Vischer N (1991) Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol 142(2–3):309–320
Zhang Y, Rowland S, King G, Braswell E, Rothfield L (1998) The relationship between hetero-oligomer formation and function of the topological specificity domain of the Escherichia coli MinE protein. Mol Microbiol 30(2):265–273
Zhou H (2009) Crowding Effects of Membrane Proteins. J Phys Chem B 113(23):7995–8005
Zhou H, Lutkenhaus J (2004) The switch I and II regions of MinD are required for binding and activating MinC. J Bacteriol 186(5):1546–1555
Zhou H, Schulze R, Cox S, Saez C, Hu Z, Lutkenhaus J (2005) Analysis of MinD mutations reveals residues required for MinE stimulation of the MinD ATPase and residues required for MinC interaction. J Bacteriol 187(2):629–638
Zimmerman S, Trach S (1991) Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol 222(3):599–620
Acknowledgments
We greatly appreciate Martin Robert, Koichi Takahashi and Alexander Popov for fruitful discussions and the critical reading of the manuscript. We thank Noam Agmon, Soohyung Park, Gleb Oshanin, Douglas Ridgway and Douglas Murray for useful discussions. We appreciate the constructive comments from the anonymous reviewers and thank the contributors and maintainers of the Gentoo Linux distribution. This work is supported by the Monbukagakusho Scholarship from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Core Research for Evolutional Science and Technology (CREST) program by the Japan Science and Technology Agency (JST) and by the research funds from the Yamagata prefectural government and Tsuruoka City, Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Arjunan, S.N.V., Tomita, M. A new multicompartmental reaction-diffusion modeling method links transient membrane attachment of E. coli MinE to E-ring formation. Syst Synth Biol 4, 35–53 (2010). https://doi.org/10.1007/s11693-009-9047-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11693-009-9047-2