Abstract
Bioinformatics is a fast-growing, diverse research field that has recently gained much public attention. Even though there are several attempts to understand the field of bioinformatics by bibliometric analysis, the proposed approach in this paper is the first attempt at applying text mining techniques to a large set of full-text articles to detect the knowledge structure of the field. To this end, we use PubMed Central full-text articles for bibliometric analysis instead of relying on citation data provided in Web of Science. In particular, we develop text mining routines to build a custom-made citation database as a result of mining full-text. We present several interesting findings in this study. First, the majority of the papers published in the field of bioinformatics are not cited by others (63 % of papers received less than two citations). Second, there is a linear, consistent increase in the number of publications. Particularly year 2003 is the turning point in terms of publication growth. Third, most researches of bioinformatics are driven by USA-based institutes followed by European institutes. Fourth, the results of topic modeling and word co-occurrence analysis reveal that major topics focus more on biological aspects than on computational aspects of bioinformatics. However, the top 10 ranked articles identified by PageRank are more related to computational aspects. Fifth, visualization of author co-citation analysis indicates that researchers in molecular biology or genomics play a key role in connecting sub-disciplines of bioinformatics.
Similar content being viewed by others
Introduction
For the recent past decades, bioinformatics, sparked by the Human Genome Initiative in 1989, has grown into the cross-disciplinary field and proliferated into new areas of life sciences (Brusic 2007). The field has been characterized as an emerging discipline driven by the needs of biologists to make use of the vast amounts of data that are constantly being accumulated in genomic, proteomics and functional genomics research (Luscombe et al. 2009). Since the field of bioinformatics has been actively expanded, it has become ever more vital to understand its current structure. The knowledge structure of and the trends in the bioinformatics field have been studied with several different approaches (Patra and Mishra 2006; Bansard et al. 2007; Glänzel et al. 2009). A majority of studies employed bibliometric analyses which is primarily used in information science. This method utilizes quantitative analysis and statistics to describe patterns of scholarly communication within a given field or body of literature (Osareh 1996).
Conventionally, the body of literature used in previous bibliometric analyses has been defined by either: (1) selecting a narrow body of literature, or (2) by searching numerous journals on a narrowly defined topic. These approaches may not accurately reflect the complete body of bioinformatics literature due to the evolving, multi-disciplinary nature of the field. Frequently new sub-domains appear and research is often published in non-bioinformatics journals. In this paper, we explore a new approach to detect the knowledge structure of bioinformatics by mining full-text articles.
The main goal of this paper is to identify the scholarly landscape of bioinformatics by analyzing full-text PubMed Central articles. To our best knowledge, none of the previous studies fully utilized text mining techniques to full-text articles for bibliometric analysis. Unlike previous studies, we analyze the core literatures from PubMed Central with various text mining techniques such as topic modeling, word co-occurrence, and named entity recognition. Even though some of studies applied text mining techniques to bibliographic data of bioinformatics (Bansard et al. 2007; Perez-Iratxeta et al. 2007), their primary focuses were not on studying the structure of and trends in bioinformatics. In addition, apart from previous studies, we create a bioinformatics-specific citation database from PubMed Central data collections and conduct citation analysis based on this custom-made database. Previous studies rely heavily on citation data provided by the Thomson Reuters’ Web of Science database for mapping the bioinformatics field. However, several concerns of studying citation impact by Web of Science arise. First, it is limited to citations from the list of journals provided in Web of Science. Butler (2006) found that the fields of chemistry, biology, physics, and medicine have only about 69.3–84.6 % of the publications found in Web of Science. Second, it has poor aggregation of minor variations of the same title and author. Belew (2005) found that only 60 % of Web of Science was listed as unique entries in about 4,000 publications used in the experiments, which indicates a significant duplicate rate.
These findings may indicate that Web of Science can’t be the only source for bibliometric analysis, and it is time to look into alternatives. These concerns led us to explore mining full-text articles for citation analysis. In this study, we conducted various text analysis as well as bibliometric analysis based on mining results. These analyses include word co-occurrence analysis, detection of country and institute with a Named Entity Recognition (NER) technique, topic modeling, publication productivity analysis, PageRank-based ranking of articles on the citation network, and visualization of the author co-citation network.
The main contributions of this paper are two-fold: First, this study maps out the current knowledge structure of the field to diagnose the maturity of the bioinformatics field and the possible direction of the field by mining the PubMed Central full-text. Second, we employed various advanced text mining techniques to analyze bibliographic data in addition to citation analysis.
Related work
There are several studies that applied bibliometric analysis to the field of bioinformatics. Glänzel et al. (2009) analyzed the core literature in bioinformatics with bibliometric analysis such as co-author citation analysis, national publication activity, citation impact etc. Huang et al. (2012) examined the citation patterns in bioinformatics journals by normalizing the journal impact factor provided in Journal Citation Report (JCR). Bansard et al. (2007) compared the bioinformatics and medical informatics literature to identify trends that are shared among both research fields to derive benefits from potential collaborative initiatives for their future. The field of bioinformatics was also studied by the relationship between active members of conferences such as conference organizers, keynote speakers, etc. for scholarly events and the representative of scholars’ prominence (Jeong et al. 2009). Perez-Iratxeta et al. (2007) performed a meta-analysis of abstracts published in MedLine and abstracts of NIH-funded project grants to determine the growth and spread of computational approaches across the various subfields of biomedicine during the past 30 years. Chen et al. (2010) introduced a multiple-perspective co-citation analysis technique to explore the structure and dynamics of co-citation networks. They combined network visualization, spectral clustering, automatic cluster labeling, and text summarization to analyze co-citation data. A major difference between their approach and the presented study is that we apply text mining techniques to a large size of full-text articles and automate citation analysis. Janssens et al. (2007) conducted a study to analyze the domain based on text mining and bibliometrics aided techniques, and aimed at improving classification of literature through the combination of linguistic and bibliometric tools. Ibáñez et al. (2009) developed a supervised learning technique to predict the possibility of a journal having a tool capable of predicting the citation count of an article within the first few years after publication would pave the way for new assessment systems. Manoharan et al. (2011) conduct bibliometric analysis of the bioinformatics field based on Thompson’s Web of Science database for a period from 2000 to 2010, aiming at evaluate the publication frequency, country, individual productivity and collaborative in this field.
Methodology
Data collection
Journal selection
Since bioinformatics is a highly interdisciplinary field, journals that contribute to bioinformatics tend to be cross-disciplinary. We select 47 bioinformatics journals that are found in PubMed Central (Table 1). The selection criteria were originally provided by Huang et al. (2011). We adopted most of the journals in their study and referred a few more sources.
Out of 47 journals, Web of Science indexes 34 journals (72 % coverage). We downloaded all available articles published in those 47 journals from PubMed Central from 2000 to early 2010. The total number of fulltext articles downloaded is 20,869. We wrote an XML parser in Java to spot elements of interest such as title, abstract, and references. Those extracted elements were stored in a relational citation database for analysis.
Procedure
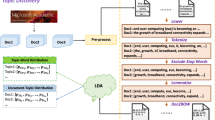
In this section, we describe the overall procedure of the proposed approach to detecting the knowledge structure of bioinformatics. First, we parse PubMed Central full-text articles to collect information elements needed for our study. Second, we build relational databases (a citation database and a text database) to store those elements. In the citation database, we create three tables such as a reference, a citation relation, and an author tables to store citation related information. In the text database, we create a full-text and an abstract tables. After the database is built, we conduct citation and text analysis. For text analysis, we employ text mining techniques such as word co-occurrence, MeSH term frequency, topic modeling, and detection of named entities. For citation analysis, we use PageRank to identify important articles and conduct bibliometric analysis for author productivity, national impact, etc. In addition, we conduct author co-citation analysis based on first author-based co-citation counts. Figure 1 illustrates the overall procedure of our approach. The details of each procedure are provided in the subsequent sections.
Word co-occurrence analysis and MeSH term frequency
To identify important concepts or themes discussed in bioinformatics, we adopt two techniques: (1) word co-occurrence and (2) MeSH term frequency. The underlying assumption and the compelling reason for adopting word co-occurrence are that words co-occurring more frequently tend to be related and show semantic connectivity of concepts. We count word co-occurrence for every pair of words in the collected datasets after filtering out a number of stop-words to come up with a total of n meaningful terms from full-text articles. We also make use of occurrences of meta-data such as MeSH to capture most frequently mentioned MeSH terms in the given datasets.
The most widely used measure of co-occurrence is mutual information (MI), a measure of the adjacent co-occurrence of words by Church and Hanks (1990). We adopt the log-likelihood ratio (LLR), a refinement of Pearson’s Chi-square test, proposed in Dunning (1993). According to Dunning, LLR is more appropriate than MI in the treatment of a mixture of high-frequency bigrams and low-frequency bigrams. The measure of the co-occurrence of u and w j is as follows:
where k 1, is the frequency with which w i occurs and is followed by w j , and n 1 is the frequency of w j , and k 2 is the frequency with which w i occurs and is followed by words other than w j , and n 2 is the frequency of words other than w i .
Another way of identifying important concepts in bioinformatics is to use MeSH terms assigned to articles. MeSH terms were used to analyze bioinformatics literatures in previous studies (Patra and Mishra 2006; Glänzel et al. 2009). Since PubMed Central articles do not contain MeSH terms, we have to map PubMed articles from PubMed Central by PubMed id to retrieve PubMed articles and then parse them in XML to count MeSH term frequency.
Detecting organization and country with NER
As part of bibliometric analysis, we are interested in the publication activity and citation impact by country and institute. Since there are no specific data fields in PubMed Central datasets for country and affiliation and many articles do not have a data field for organization, we apply the Named Entity Recognition (NER) technique to identify country and organization associated with authors in a full-text paper. To this end, we use Learning Based Java (LBJ), a perceptron-based Named Entity Recognition (NER) system (Ratinov and Roth 2009). LBJ proves to be an excellent NER technique for our study in that LBJ achieved 90.8 F1 score on the CoNLL-2003 NER shared task at the CoNLL competition in 2003, which was the best reported result of the NER shared task. The sample input and output of our NER task is given in Fig. 2.
Topic modeling for bioinformatics by LDA
We explore the salient topics in core literatures of bioinformatics. We use Latent Dirichlet Allocation (LDA) for topic model generation (Blei et al. 2003). LDA, a statistical learning algorithm, is a generative model that enables to account for a set of hidden topic structures by using the observed documents to infer the hidden structures embedded in the collection. The underlying intuition of LDA is that documents exhibit multiple topics. In LDA, each group is described as a random mixture over latent topics where each topic is a discrete distribution over the vocabulary of the collection. The generative process for a document collection D under the LDA model is as follows: For k = 1…K: (a) \( \varphi^{(k)} \sim {\text{Dirichlet}}(\beta ) \) and for each document \( d \in D: \) (a) \( \theta_{d} \sim {\text{Dirichlet}}(\alpha ) \)and (b) For each word \( w_{i} \in d:1)z_{i} \sim {\text{Discrete}}(\theta_{d} ) \) and 2) \( w_{i} \sim {\text{Disctete}}(\varphi^{{(z_{i} )}} ) \) where K is the number of latent topics in the collection, φ (k) is a discrete probability distribution over a fixed vocabulary that represents the kth topic distribution, θ d is a document-specific distribution over the available topics, z i is the topic index for word w i , and α and β are hyper-parameters for the symmetric Dirichlet distributions that the discrete distributions are drawn from. The generative process described above results in the following joint distribution:
Each θ d is a low-dimensional representation of a document in a topic space, each z i represents which topic generated the word instance w i , and each φ (k) represents a K × V matrix where \( \varphi_{i,j} = p(w_{i} |z_{j} ) \). Therefore, one of the most interesting aspects of LDA is that it can learn, in an unsupervised manner, words that we would associate with certain topics, and this is expressed through the topic distributions φ. For maximum likelihood (ML) estimation of the LDA model the log-likelihood of the data is maximized with respect to the model parameters α and β which are in general the parameter of interest. Since the quantities \( p(w|\alpha ,\beta ) \) for the LDA model is not tractably computed, we use the expectation maximization procedure.
Author co-citation analysis
Author Co-citation analysis (ACA) has been a compelling bibliometric method in Information Science. ACA uses authors as the units of analysis and the co-citations of pairs of authors as the variable that indicates their dissimilarity from each other. The underlying assumption of ACA is that the more two authors are cited together, the closer the relationship between them (White and Griffith 1981).
In our study, we fully automated the ACA procedure which is one of the main contributions of this study. Most ACA studies including White and McCain (1998) select either manually or semi-automatically key journals from Web of Science, select top N authors ranked by citation counts, and visualize a field through a representative slice of its literature. In visualization, ACA studies select at most 300 authors due to the limitation of the software used if the study employs Multi-dimensional Scaling (MDS). Unlike previous studies, we developed an automated, scalable procedure for ACA to overcome the problems of existing approaches. The procedures include calculating co-citation pairs from the entire author list, constructing co-citation count table in a relational database, and integrating several visualization tools such as Gephi and Prefuse via APIs.
Ranking important articles by PageRank
We use PageRank to identify important articles in bioinformatics. We apply PageRank for spotting important articles in the citation network since PageRank can nicely work with the citation network. PageRank provides an effective way to evaluate the relative importance of publications beyond mere citation counts (Ding et al. 2009). In Bibliometrics, the number of citations is used to measure the impact of scientific publications. However, there is a critical issue with this measurement that it does not reflect the importance of the citing papers. That is, a citation from a mediocre paper has the same weight as a citation from a highly cited work (Maslov and Redner 2008). The PageRank algorithm can overcome this shortcoming in that it gives higher weights to the publications that are highly cited and also to papers cited by a few highly cited papers. PageRank is adopted as a complementary method to citation analysis, which allows us to identify publications referenced by highly cited articles.
Results and discussion
In this section, we report the results of mining bioinformatics literatures in terms of (1) text analysis and (2) link analysis.
Word co-occurrence analysis
To calculate word co-occurrence from 20,869 bioinformatics related articles, we filter out common words that are normally used in Information Retrieval. It consists of 450 words such as ‘and’, ‘or’, ‘which’, etc. In addition, co-occurrence of words is calculated from abstracts of the data collection. Table 2 shows the list of keywords identified with word co-occurrence. The importance of word co-occurrence is measured by LLR. Higher LLR scores mean pairs of terms with the more interestingly connected terms. In this usage, the LLR has proven very useful for discriminating pairs of features that have interesting degrees of co-occurrence.
The results of word co-occurrence analysis indicate that highly co-occurred terms can be by and large classified into two categories: biology and computer. Biological terms like gene, genome are most dominant concepts in bioinformatics datasets from PubMed Central. Computer related terms like data, algorithm, and database also are co-occurred with high LLR scores.
In addition, individual word pairs with high LLR scores are presented in Table 3. The word pair “Gene expression” is ranked top and its LLR score is two times bigger than the second ranked pair “amino acid”. As shown in Table 3, top 12 word pairs are all related to molecular biology.
Frequency of MeSH terms
Out of 20,869 documents, there are 19,954 documents that have the corresponding MEDLINE records (95.6 % matching). In 19,954 documents, 8,412 documents have MeSH terms (42.2 %).
Table 4 shows the frequently occurred MeSH terms in bioinformatics literatures. Since there are only 42 % of full-text articles that have MeSH terms, it is not desirable to compare directly to LLR based co-occurrence. However, MeSH terms with high frequency may show us an overview of how the structure of bioinformatics looks like in terms of controlled vocabulary. Except for the top two terms (Animals and Humans), the list of MeSH terms is related to either of the topics biology and computer. A major difference between word co-occurrence with LLR and MeSH terms is that in MeSH terms, there are more computer related terms, such as algorithm and software that are highly ranked than the results of word-co-occurrence analysis. However, the majority of dominant concepts, pertinent to Computational Biology and Genomics, are the same between two approaches.
Topic modeling for bioinformatics literature
Table 5 shows the 10 topics generated with LDA, and we describe these topics briefly. Topic 1 has something to do with transcription of DNA in the Yeast, genome sequence, and gene expression regulation. A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Topic 2 is mainly related to the topic of Computational Genomics and gene prediction. In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. Topic 3 deals with Evolutionary Homologs and Caenorhabditis elegans that are a free-living, transparent nematode (roundworm) and an accelerated rate of evolution in the C. elegans lineage. Topic 4 is about embryonic stem cells and molecular cancer. Topic 5 is pertinent to data mining and Proteomics. Topic 6 describes DNA methylation and sequencing. Topic 7 is relevant to pathway and gene regulation. Topic 8 is related to System Biology and protein network. Topic 9 is associated with Biogenesis and cellular function. Topic 10 is related to RNS inference and Drosphilia Genome.
These results of topic modeling indicate that the majority topics are related to biological aspects rather than computational aspects of bioinformatics. Only two topics (Topic 2 and Topic 5) focus more on computational aspects. The rest of the topics are related to gene or DNA sequencing, System Biology, and protein network to some extent. Topic 4 and 5 are associated with some special topics such as Evolutionary Homologs and embryonic stem cells. These results conform to the results of word co-occurrence analysis.
Research productivity analysis
In this section, we present the results of research productivity analysis as a result of mining PubMed Central full-text articles. As mentioned in the “Methodology” section, we examine changes in the number of citations over time, author productivity, publication growth over time, and research productivity by institutes and countries. As illustrated in Fig. 3, we observe that the relationship between the number of papers and the number of citation a paper receives follows Zipf’s law. Among 740,353 papers (drawn from 20,869 papers and its citations), 285,439 citations receive 1 citation, 182,548 papers received 2 citations, and 138,090 papers receive 3 citations. Figure 3 shows the relationship between a paper and the number of citations it receives.
The skewness issue of scientific publications has been reported by several researchers (Seglen 1992; van Raan 2006; Stringer et al. 2010; Albarrán and Ruiz-Castillo 2011; Franceschet 2011). Franceschet reported that 21 % of journal papers and 56 % of conference papers received zero citation in the Computer Science related conferences and journals (Franceschet 2011). Alb Albarrán and Ruiz-Castillo (2011) collected 7 million articles from 22 research fields and observed that 74.7 % of the dataset follows the power law distribution. In our case, we have a much higher rate of receiving zero citation than in those related works. It may be attributed to the characteristics of our data collection that citation counts are limited to 20,869 full-text articles and their references.
Author productivity
Authors with single publication were predominant (77.7 %) which is higher than the predicted percentage (73.58 %) calculated by Lotka’s law. Patra and Mishra’s study show the similar observation that there are 73.58 % of authors with single publication in bioinformatics (Patra and Mishra 2006). Figure 4 shows the relationship between the number of authors and the citations an author receives.
Based on this result together with the evidence of the fast growth rate reported in Fig. 5, we assume that the number of researchers entering the field of bioinformatics keeps increasing. The field is still in the growing phase and has not reached maturity.
Publication productivity by year
We examined the publication productivity by year. Out of the total number of full-text articles (20,869), Fig. 5 shows that there is a dramatical increase in publication in 2003 and onward. Note that the data for year 2010 is not complete since we collected the data in February 2011. While this may not represent the whole picture of bioinformatics in terms of publication productivity, it at least indicates that bioinformatics is a fast growing field.
This stiff increase in the number of publications from 2003 and on is also observed in Web of Science bioinformatics data. The number of articles is calculated by summing up the articles assigned to subject areas of 34 overlapped journals with our data collection by Web of Science.
Important papers by PageRank
The results of article ranking by PageRank are shown in Table 6 along with the title and the journal title. The first top two articles are written by Altschul et al. Both articles are related to BLAST algorithms (Altschul et al. 1990, 1997). The third ranked article is by Ashburner et al. (2000) which introduces the Gene Ontology (GO) tool. Among top 10 articles, three articles were published in Nucleic Acids Research, and nine articles are journal articles and one is a book.
One interesting observation is that most of highly ranked articles focus more on computational aspects of bioinformatics rather than biological aspects. This shows the different results from topic modeling where biological aspects of bioinformatics are dominant. This is further scrutinized in discussion of Author Co-citation Analysis later in this paper.
Research productivity by country and institute
To understand the research productivity by country and institute respectively, we first extract country and institute names by NER. The 30 most active countries in the period 2000–2010 have been selected. We count country names that occur in the affiliation address field. Figure 6 shows that USA is most productive followed by UK.
Among top 10 productive countries are USA, European, Asian countries.
Table 7 shows top 20 universities in terms of the research productivity by institute. University of California is ranked first because multiple campuses in California are counted as University of California together. Therefore, the number one institute as a single body is Harvard followed by Stanford.
Except for two universities (University College London and University of Toronto), all universities are based in USA.
Author co-citation analysis
We conducted author co-citation analysis. We calculated all co-citation pairs. The number of pairs is 339,121,666. This number is based on the first author co-citation count. Since it is too big, it takes too long to calculate co-citation count even on the high end computer. To overcome this big data issue, we used the MapReduce technique that supports data intensive distributed applications. The MapReduce technique was proposed by Google as part of their distributed computing model for processing large data sets. MapReduce consists of two operations: map and reduction operations. The mapping operation is independent of the others. All maps can be performed in parallel. Similarly, a set of reduce operations can perform the reduction phase—provided all outputs of the map operation that share the same key are presented to the same reduce operation at the same time. We built our co-citation technique based on Apache Hadoop developed in Java (http://hadoop.apache.org/). Once we calculated the co-citation count for all author pairs, we order the pairs in terms of frequency. We selected the top 200 authors by rank of the co-citation count. With these 200 authors, we built a co-citation matrix and applied the PFNET scaling technique to the matrix. Our ACA technique is the bottom up approach whereas existing ACA techniques use the top down approach. The top down approach means that the top N highly cited authors are selected first and then compute the pair counts between the author(s) of highly cited papers and author(s) of the citing papers. Instead, our approach first computes all possible pairs of citing authors and cited authors. The pairs of authors to be counted are enormous, and it can only be handled distributed computing techniques like MapReduce. The detailed description of the author co-citation analysis technique is provided in our forthcoming paper (Song and Chung 2013).
In terms of visualization, we employed Gephi’s visualization technique (http://gephi.org/). Figure 7 illustrates the author co-citation network with Gephi. We used the betweenness centrality to calculate the node distance. Betweenness centrality is a measure based on the number of shortest paths between any two nodes that pass through a particular node. Nodes around the edge of the network tend to have a low betweenness centrality whereas a high betweenness centrality indicates that the individual is connecting various different parts of the network together. In Fig. 7, bigger nodes, meaning a higher centrality, indicates that researchers on the bigger nodes play a key role in connecting different sub-disciplines of bioinformatics. For instance, Teusink is a scholar in System bioinformatics who collaborates with researchers interested in biological networks. Cox is a molecular biologist who is particularly interested in genetic rearrangements. Both Teusink and Cox have a major in Biochemistry. In the lower left corner, there appear researchers like Eggeling in Proteomics and H. Berman in biological databases. Eggeling is in the Biotechnology department and Berman is in the department of Molecular Bioscience.
Visualization results indicate that biologists or biochemistry scientists receive higher recognition and have higher visibility than computation oriented researchers in the field of bioinformatics. This result is in aligned with the results of word co-occurrence analysis, but not with the results of PageRank based citation ranking on the citation network. We further investigated whether the difference is attributed to the fact that PageRank is applied to the citation network not the co-citation network. To this end, we built the co-citation network with the author pairs whose co-citation count is greater than 5. The total number of pairs is 997,415. We applied PageRank with the damping factor set to 0.15 which is the same for PageRank on the citation network.
Table 8 shows the results of top 5 important authors in the co-citation network ranked by PageRank. This result coincides with the result of ACA, and it also reveals that the important authors are leaning more toward biological aspects than computational aspects of bioinformatics. This result also confirms that the findings reported by Ding and her colleagues that the ranking of authors in the author co-citation network are heavily influenced by whom the author is co-cited with (Ding et al. 2009). In other words, if an author is co-cited with important authors, which means the author who has high co-citation counts, a high PageRank score is assigned to the author. As shown in Table 8, authors ranked in the top 5 are co-cited with most of important authors. This is in turn confirmed by Fig. 7. For instance, the number one ranked author, Palsson, is co-cited with Reed, Eggeling and Teusink who are regarded as important authors in the co-citation network.
Conclusion
The field of bioinformatics is considered to be a fast-growing, interdisciplinary field with the vast public attention starting from early 2000. In this paper, we explore the knowledge structure of and trends in bioinformatics by applying text mining techniques to PubMed Central full-text articles. Besides several core journals, important periodicals in molecular biology as well as the multidisciplinary journals such as Science and Nature proved to be the most important publication channels. Although we focused on the bioinformatics core literature, our study has confirmed findings by other recent studies concerning publication patterns. There are several interesting findings reported in this paper. First, the majority of the papers were not cited by others (83 % of papers received zero citation). Second, we observed that there is a linear, consistent increase in the number of publications. Particularly year 2003 is the turning point. Third, most researches of bioinformatics are driven by USA-based institutes followed by European institutes. Fourth, the results of topic modeling and word co-occurrence analysis reveal that major topics are closer to biological aspects than computational aspects of bioinformatics. But top 10 ranked articles identified by PageRank are more related to computational aspects. Fifth, visualization of ACA indicates that researchers in molecular biology or genomics play a key role in connecting sub-disciplines of bioinformatics. This visualization result is confirmed by important authors identified by PageRank in the author co-citation network.
The contributions of our paper are three-folds: (1) it is the first attempt to fully utilize text mining techniques to understand the knowledge structure of a field. (2) We chose PubMed Central full-text data and automated citation analysis based on the PubMed Central data. 3) we conducted comprehensive content as well as citation analysis. As a follow-up study, we plan to compare the results of our approach to PubMed Central data with the traditional approach which is based on Web of Science citation data. We are also interested in exploring new ways of utilizing citation data to discover new hypothesis generation in bioinformatics.
References
Albarrán, P., & Ruiz-Castillo, J. (2011). References made and citations received by scientific articles. Journal of the American Society for Information Science and Technology, 62(1), 40–49.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, M., et al. (2000). Gene Ontology: tool for the unification of biology. Nature Genetics, 25(1), 25–29.
Bansard, J. Y., Rebholz-Schuhman, D., Cameron, G., Clark, D., van Mulligen, E., Beltrame, F., et al. (2007). Medical informatics and bioinformatics: a bibliometric study. IEEE Transactions on Information Technology in Biomedicine, 11(3), 237–243.
Belew, R.K. (2005). Scientific impact quantity and quality: Analysis of two sources of bibliographic data. arXiv:cs.IR/0504036 v1. pp. 1–12.
Blei, D., Ng, A., & Jordan, M. (2003). Latent Dirichlet allocation. Journal of Machine Learning Research, 3, 993–1022.
Brusic, V. (2007). The growth of bioinformatics. Briefings in Bioinformatics., 8(2), 69–70.
Butler, L. (2006). RQF Pilot Study Project—History and Political Science Methodology for Citation Analysis, November 2006. http://www.chass.org.au/papers/PAP20061102LB.php. Accessed 14 Oct 2012.
Chen, C., Ibekwe-SanJuan, F., & Hou, J. (2010). The structure and dynamics of cocitation clusters: A multiple-perspective cocitation analysis. Journal of American Society for Information Science, 61(7), 1386–1409.
Church, K., & Hanks, P. (1990). Word association norms, mutual information and lexicography. Computational Linguistics, 16(1), 22–29.
Ding, Y., Yan, E., Frazho, A., & Caverlee, J. (2009). PageRank for ranking authors in co-citation networks. Journal of the American Society for Information Science and Technology, 60(11), 2229–2243.
Dunning, T. (1993). Accurate methods for the statistics of surprise and coincidence. Computational Linguistics, 19(1), 61–74.
Franceschet, M. (2011). The skewness of computer science. Information Processing and Management, 47(1), 117–124.
Glänzel, W., Janssens, F., & Thijs, B. (2009). A comparative analysis of publication activity and citation impact based on the core literature in bioinformatics. Scientometrics, 79(1), 109–129.
Huang, H., Andrews, J., & Tang, J. (2011). Citation characterization and impact normalization in bioinformatics journals. Journal of the American Society of Information Science and Technology, 63(3), 490–497.
Ibáñez, A., Larrañaga, P., & Bielza, C. (2009). Predicting citation count of Bioinformatics papers within four years of publication. Bioinformatics, 25(24), 3303–3309.
Janssens, F., Glänzel, W., & De Moor, B. (2007). Dynamic hybrid clustering of bioinformatics by incorporating text mining and citation analysis. In Proceedings of the 13th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD 07), pp. 360–369.
Jeong, S., Lee, S., & Kim, H. G. (2009). Are you an invited speaker? A bibliometric analysis of elite groups for scholarly events in bioinformatics. Journal of the American Society for Information Science and Technology, 60(6), 1118–1131.
Luscombe, N. M., Greenbaum, D, & Gerstein, M. (2001). What is bioinformatics? A proposed definition and overview of the field. Methods of Information in Medicine, 40, 346–58.
Manoharan, A., Kanagavel, B., Muthuchidambaram, A., Kumaravel, J.P.S. (2011) Bioinformatics Research – An Informetric View. In 2011 International Conference on Information Communication and Management (IPCSIT) vol.16.
Maslov, S., & Redner, S. (2008). Promise and pitfalls of extending Google’s PageRank algorithm to citation networks. Journal of Neuroscience, 28(44), 11103–11105.
Osareh, F. (1996). Bibliometrics, citation analysis and co-citation analysis: A review of literature I. Libri, 46(3), 149–158.
Patra, S. K., & Mishra, S. (2006). Bibliometric study of bioinformatics literature. Scientometrics, 67(3), 477–489.
Perez-Iratxeta, C., Andrade-Navarro, M. A., & Wren, J. D. (2007). Evolving research trends in bioinformatics. Briefings in Bioinformatics, 8(2), 88–95.
Ratinov, L., & Roth D. (2009). Design challenges and misconceptions in named entity recognition. In Proceedings of the Thirteenth Conference on Computational Natural Language Learning (CoNLL 09), pp. 147–155.
Seglen, P. O. (1992). The skewness of science. Journal of the American Society for Information Science, 43(9), 628–638.
Song, M., & Chung, Y.K. (2013). Mining citation data for automatic author co-citation analysis, to be submitted to Information Processing and Management.
Stringer, M. J., Sales-Pardo, M., & Nunes Amaral, L. A. (2010). Statistical validation of a global model for the distribution of the ultimate number of citations accrued by papers published in a scientific journal. Journal of the American Society for Information Science and Technology, 61(7), 1377–1385.
van Raan, A. F. J. (2006). Statistical properties of bibliometric indicators: Research group indicator distributions and correlations. Journal of the American Society for Information Science and Technology, 57(3), 408–430.
White, H. D., & Griffith, B. C. (1981). Author cocitation: A literature measure of intellectual structure. Journal of American Society for Information Science, 32(3), 163–171.
White, H. D., & McCain, K. W. (1998). Visualizing a discipline: An author co-citation analysis of information science, 1972–1995. Journal of the American Society for Information Science, 49(4), 327–355.
Acknowledgments
We give a special think to Ying Ding for her invaluable comments on the manuscript to improve the quality of the paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, M., Kim, S.Y. Detecting the knowledge structure of bioinformatics by mining full-text collections. Scientometrics 96, 183–201 (2013). https://doi.org/10.1007/s11192-012-0900-9
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-012-0900-9