Abstract
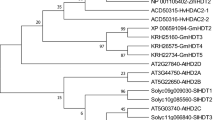
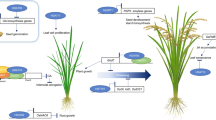
Epigenetic regulation of gene expression plays an important role in various aspects of eukaryotic development and is associated with modifications of chromatin structure. These are accomplished, in part, through the reversible process of histone acetylation/deacetylation, catalyzed by histone acetyltransferases (HATs), and histone deacetylases (HDACs), respectively. Eukaryotic HDACs are grouped in three major families, RPD3/HDA1 (thereafter cited as HDA1), SIR2 and the plant-specific HD2. Histone deacetylase genes have been studied in Arabidopsis and rice, but little is known about these genes from important crop plants. In this work, cDNAs encoding members of the HDA1 family and representing all four classes, Class I, Class II, Class III, and Class IV, were isolated and characterized from barley (Hordeum vulgare), a cereal crop of high agronomic importance. Expression analysis of the barley HDA1 family genes, HvHDAC1-I-1, HvHDAC1-I-3, HvHDAC1-II-1, HvHDAC1-III-1, and HvHDAC1-IV-1 demonstrated that they are expressed in all tissues and seed developmental stages examined. Differences in transcript abundance both in vegetative and reproductive tissues were observed among the different genes suggesting functional diversification of the HDA1 members. Differential expression was also evidenced for some of the HDA1 genes in two barley cultivars differing in various characteristics, such as seed size and resistance to stress, implying a possible association of these genes with different traits. Furthermore, the HDA1 genes were found to respond to the stress-related hormone jasmonic acid (JA), suggesting an association of these genes with barley responsiveness to biotic and abiotic stress. The expression pattern of the HDA1 genes suggests possible roles in the epigenetic regulation of barley development and stress response.





Similar content being viewed by others
References
Ausfatz W, Mette MF, Van Der Winden J, Matzke M, Matzke AJ (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double- stranded RNA. EMBO J 21:6832–6841
Chen ZJ, Tian L (2007) Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 1769:295–307
Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12:133–139
Chinnusamy V, Gong Z, Zhu JK (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol 50:1187–1195
Demetriou K, Kapazoglou A, Tondelli A, Francia E, Stanca MA, Bladenopoulos K, Tsaftaris AS (2009) Epigenetic chromatin modifiers in barley: I. Cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiol Plant 136:358–368
Fu W, Wu K, Duan J (2007) Sequence and expression analysis of histone deacetylases in rice. Biochem Biophys Res Comm 356:843–850
Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128:635–638
Hassig CA, Tong JK, Fleischer TC et al (1998) A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA 95:3519–3524
Hollender C, Zhong L (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50:875–885
Kadosh D, Struhl K (1998) Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev 12:797–805
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5:150–163
Lippman Z, May B, Yordan C, Singer T, Martienssen R (2003) Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1:420–428
Loidl P (2004) A plant dialect of the histone language. Trends Plant Sci 9:84–90
Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312:1520–1523
Lusser A, Kolle D, Loidl P (2001) Histone acetylation: lesson from the plant kingdom. Trends Plant Sci 6:59–65
Pandey R, Muller A, Napoli CA et al (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30:5036–5055
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36
Polidoros AN, Pasentsis K, Tsaftaris AS (2006) Rolling circle amplification-RACE: a method for simultaneous isolation of 5′ and 3′ cDNA ends from amplified cDNA templates. Biotechniques 4:35–36
Probst AV, Fragard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, Vaucheret H, Scheid OM (2004) Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16:1021–1034
Rossi V, Locatelli S, Lanzanova C, Boniotti MB, Varotto S, Pipal A, Goralik-Schramel M, Lusser A, Gatz C, Gutierrez C, Motto M (2003) A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol Biol 51:401–413
Rossi V, Locatelli S, Varotto S, Donn G, Pirona R, Henderson DA, Hartings H, Motto M (2007) Maize histone deacetylase hda101 is involved in plant development, gene transcription and sequence specific modulation of histone modification of genes and repeats. Plant Cell 19:1145–62
Saitou N, Nei M (1987) The neighbor-joining method:a new method for reconstructing phylogenetic trees. Mol Biol E 4:406–425
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46:124–133
Tanaka M, Kikuchi A, Kamada H (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146:149–161
Taunton J, Hassing CA, Schreiber SL (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408–411
Tian L, Chen ZL (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA 98:200–205
Tian L, Fong MP, Wang J, Wei N, Jiang H, Doerge RW, Chen ZJ (2005) Reversible histone acetylation and deacetylation mediate genome-wide, promote-dependent and locus-specific changes in gene expression during plant development. Genetics 169:337–345
Thomson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Close TJ (2007) Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ 30:410–421
Wu K, Tian L, Malik K, Brown D, Miki B (2000) Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J 22:19–27
Wu K, Zhang L, Zhou C, Yu CW, Chaikam V (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59:225–234
Wu JI, Lessard J, Crabtree GR (2009) Understanding the words of chromatin regulation. Cell 136:200–206
Xu C, Liu C, Wang Y et al (2005) Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Pro Natl Acad Sci USA 102:14469–14474
Yang XJ, Seto E (2003) Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev 13:143–153
Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Yang Z, Brown D, Miki B, Wu K (2004) Expression and function of HD2-type histone deacetylase in Arabidopsis development. Plant J 38:715–724
Zhou C, Zhang L, Duan J, Miki B, Wu K (2005) HISTONE DEACETYLASE 19 is involved in jasmonic acid and ethylene signalling of pathogen response in Arabidopsis. Plant Cell 17:1196–1204
Acknowledgments
This work was supported by a PENED grant (Ο3ΕΔ402/2003). Continuous support for the Institute of Agrobiotechnology/CERTH from the General Secretariat of Research and Technology of Greece is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kyproula Demetriou and Aliki Kapazoglou contributed equally to this work.
Rights and permissions
About this article
Cite this article
Demetriou, K., Kapazoglou, A., Bladenopoulos, K. et al. Epigenetic Chromatin Modifiers in Barley: II. Characterization and Expression Analysis of the HDA1 Family of Barley Histone Deacetylases During Development and in Response to Jasmonic Acid. Plant Mol Biol Rep 28, 9–21 (2010). https://doi.org/10.1007/s11105-009-0121-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-009-0121-4