Abstract
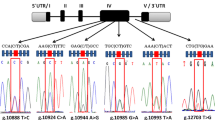
Leucine aminopeptidase 3 (LAP3) is an aminopeptidase which catalyses the removal of N-terminal amino acids and is involved in protein maturation and degradation. In this study, we detected the polymorphisms of LAP3 gene by polymerase chain reaction-single-strand conformation polymorphism (PCR-SSCP) and DNA sequencing methods in 916 individuals from three Chinese cattle breeds including Chinese Holstein, Luxi Yellow and Bohai Black. One novel single nucleotide polymorphism (SNP) (g.24564G>A ss196003366) and four previously deposited SNPs in the GenBank database (g.24794T>G, g.24803T>C, g.24846T>C, g.25415T>C) were detected. Three of the SNPs (g.24794T>G, g.24803T>C, g.24846T>C) were firstly found to be linked completely and regarded as a SNP g.24794M>N by PCR-SSCP and DNA sequencing in the tested breeds. The allelic frequencies and genetic indices of the SNPs were different in three Chinese cattle populations. The SNPs and their genetic effects on milk production traits in Chinese Holsteins were evaluated. Least squares analysis showed that cows with genotype MM had higher fat percentage and protein percentage than genotype NN (P < 0.05); and the cows with g.25415T>C-CC genotype had higher protein rate than ones with TT genotype (P < 0.05). In addition, eight haplotypes and 23 combined genotypes were identified based on the nine genotypes and the association between combined genotypes and milk production traits were analyzed. Statistic results showed that the cows with genotype combination MAT/MGC have higher protein and fat rate and lower SCS. Our finding demonstrated that the LAP3 gene possibly contributed to conducting association analysis and can be used as molecular marker in milk production traits and other performance for animal breeding.



Similar content being viewed by others
References
Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, Cambisano N, Mni M, Reid S, Simon P (2002) Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res 12:222–231
Cohen-Zinder M, Seroussi E, Larkin DM, Loor JJ, Wind AE, Lee JH, Drackley JK, Band MR, Hernandez AG, Shani M, Lewin HA, Weller JI, Ron M (2005) Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res 15:936–944
Lü A, Hu X, Chen H, Jiang J, Zhang C, Xu H, Gao X (2010) Single nucleotide polymorphisms in bovine PRL gene and their association with milk production traits in Chinese Holsteins. Mol Biol Rep 37(1):547–551
Wayne ML, McIntyre LM (2002) Combining mapping and arraying: an approach to candidate gene identification. Proc Natl Acad Sci 99:14903–14906
Khatkar MS, Thomson PC, Tammen I, Raadsma HW (2004) Quantitative trait loci mapping in dairy cattle: review and meta-analysis. Genet Sel Evol 36:163–190
Olsen HG, Lien S, Gautier M, Nilsen H, Roseth A, Berg PR, Sundsaasen KK, Svendsen M, Meuwissen THE (2005) Mapping of a milk production quantitative trait locus to a 420-kb region on bovine chromosome 6. Genetics 169:275–283
Cuypers HT, van Loon-Klaassen LA, Egberts WT, de Jong WW, Bloemendal H (1982) The primary structure of leucine aminopeptidase from bovine eye lens. J Biol Chem 25(7):7077–7085
Matsui M, Fowler JH, Walling LL (2006) Leucine aminopeptidases: diversity in structure and function. Biol Chem 387(12):1535–1544
Kloetzel PM, Ossendorp F (2004) Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol 16(1):76–81
Tsujimoto M, Mizutanl S, Adachi H, Kimura M, Nakazato H, Tmomda Y (1992) Identification of human placental leucine aminopeptidase as oxytocinase. Arch Biochem Biophys 292:388–392
Huang J, Wang H, Wang C, Li J, Li Q, Hou M, Zhong J (2010) Single nucleotide polymorphisms, haplotypes and combined genotypes of lactoferrin gene and their associations with mastitis in Chinese Holstein cattle. Mol Biol Rep 37(1):477–483
Zhang CL, Wang YH, Chen H, Lan XY, Lei CZ (2007) Enhance the efficiency of single-strand conformation polymorphism analysis by short polyacrylamide gel and modified silver staining. Anal Biochem 365:286–287
Zhao CJ, Li N, Deng XM (2003) The establishment of method for identifying SNP genotype by CRS-PCR. Yichuan 25:327–329
Yeh FC, Yang RC, Boyle T (1999) POPGENE Version 1.31, Microsoft window-based freeware for population genetic analysis. University of Alberta and Centre for International Forestry Research, Canada
Shi YY, He L (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15:97–98
Ali AKA, Shook GE (1980) An optimum transformation for somatic cell concentration in milk. J Dairy Sci 63:487–490
Rupp R, Boichard D (1999) Genetic parameters for clinical mastitis, somatic cell score, production, udder type traits and milking ease in first lactation Holsteins. J Dairy Sci 82:2198–2204
Qiu H (2002) Modern dairy science. Agriculture Press of China, pp 1–12
Sheely PA, Riley LG, Raadsma HW, Williamson P, Wynn PC (2009) A function genomics approach to evaluate genes located in a QTL interval for milk production traits on BTA6. Anim Genet 40:492–499
Taylor A (1993) Aminopeptidases: structure and function. FASEB J 7:290–298
Noboru Y, Seiji N, Takanobu S, Atsuo I, Mitsuaki I, Tomomitsu O, Masafumi T, Hiroshi N, Shigehiko M (2000) Placenta leucine aminopeptidase/oxytocinase in maternal serum and placenta during normal pregnancy. Life Sci 15:1401–1410
Nott A, Meislin SH, Moore MJ (2003) A quantitative analysis of intron effects on mammalian gene expression. RNA 9(5):607–617
Zan LS, Zhang JL, Liu XW (2007) Association study on AGPAT6 intron3 polymorphism and milk performance of dairy cattle. Sci Agric Sin 40:1498–1503
Chen JM, Férec C, Cooper DN (2006) A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet 120:1–21
Kubiak EJ, Flisikowski K, Wicińska K (2010) A new SNP in the 3′UTR region of the bovine calpain small subunit (CAPNS1) gene. Mol Biol Rep 37:473–476
Han Q, Wang SZ, Hu G, Li H (2009) Correlation between peroxisome proliferator-activated receptor γ 5′Flanking-region haplotypes with the growth and body composition traits in chickens. Sci Agric Sin 42(10):3647–3654
Fallin D, Cohen A, Essioux L, Chumakov I, Blumenfeld M, Cohen D, Schork NJ (2001) Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer’s disease. Genome Res 11(1):143–151
Acknowledgments
This project was partly funded by the National “863” Program of China (No. 2007AA10Z169, No. 2006AA10Z1D9), National Cow Industrial Technology System Program (No. nycytx-0107, No. nyhyzx07-036-09), Well-bred Program from Shandong province (No. 2007LZ10-06), and Key Scientific and Technological Project from Shandong province (No. 2009GG20002033).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, X., Ju, Z., Wang, J. et al. Single nucleotide polymorphisms, haplotypes and combined genotypes of LAP3 gene in bovine and their association with milk production traits. Mol Biol Rep 38, 4053–4061 (2011). https://doi.org/10.1007/s11033-010-0524-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0524-1