Abstract
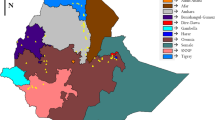
In situ population studies of wild relatives of crops are crucial for the conservation of plant genetic resources, especially in regions with high genetic diversity and a risk of local extinction. Ethiopia is the center of origin for sorghum, yet little is known about the genetic structure of extant wild populations. Using 9 Simple Sequence Repeat loci, we characterized 19 wild populations from five regions, 8 local cultivar populations from three regions, and 10 wild sorghum accessions from several African countries. To our knowledge, this is the most comprehensive study to date of in situ wild sorghum populations in Africa. Genetic diversity corrected for sample size was significantly greater in the wild populations in situ than in local cultivars or the accessions. Approximately 41 % of the genetic variation in the wild plants was partitioned among populations, indicating a high degree of differentiation and potential value for germplasm conservation, and the average number of migrants (Nm) per generation was 0.43. Cluster analyses showed that some wild populations were grouped by geographic region, whereas others were not, presumably due to long-distance seed movement. Four wild populations from disjunct regions formed a unique cluster with an Ethiopian accession of subsp. drummondii and probably represent a weedy race. STRUCTURE and other analyses detected evidence for crop-wild hybridization, consistent with previous molecular marker studies in Kenya, Mali, and Cameroon. In summary, in situ wild sorghum populations in Ethiopia harbor substantial genetic diversity and differentiation, despite their close proximity to conspecific cultivars in this crop/wild/weedy complex.



Similar content being viewed by others
References
Abu-Assar AH, Uptmoor R, Abdelmula AA, Salih M, Ordon F, Friedt W (2005) Genetic variation in sorghum germplasm from Sudan, ICRISAT, and USA assessed by simple sequence repeats (SSRs). Crop Sci 45:1636–1644
Adugna A (2012) Population genetics and ecological studies in wild sorghum [Sorghum bicolor (L.)] in Ethiopia: implications for germplasm conservation. PhD Thesis, Addis Ababa University
Adugna A, Snow AA, Sweeney P (2011) Optimization of a high throughput, cost effective and all-stage DNA extraction protocol for sorghum. J Agric Sci Tech 5:243–250
Agrama HA, Tuinstra MR (2003) Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. Afr J Biotech 2:334–340
Aldrich PR, Doebley J (1992) Restriction fragment variation in the nuclear and chloroplast genomes of cultivated and wild Sorghum bicolor. Theor Appl Genet 85:293–302
Aldrich PR, Doebley J, Schertz KF, Stec A (1992) Patterns of allozyme variation in cultivated and wild Sorghum bicolor. Theor Appl Genet 85:451–460
Ayana A, Bekele E, Bryngelsson T (2001) Genetic variation in wild sorghum (Sorghum bicolor subsp. verticilliflorum (L.) Moench) germplasm from Ethiopia assessed by random amplified polymorphic DNA (RAPD). Hereditas 132:249–254
Barnaud A, Trigueros G, McKey D, Joly H (2008) High outcrossing rates in fields with mixed sorghum landraces: how are landraces maintained? Heredity 101:445–452
Barnaud A, Deu M, Garine E, Chantereau J, Bolteu J, Koıda EO, McKey D, Joly HI (2009) A weed–crop complex in sorghum: the dynamics of genetic diversity in a traditional farming system. Am J Bot 96:1869–1879
Barrett SCH (1983) Crop mimicry in weeds. Econ Bot 37:255–282
Bhattramakki D, Dong J, Chhabra AK, Hart GE (2000) An integrated SSR and RFLP linkage map of Sorghum bicolor (L.) Moench. Genome 43:988–1002
Bohonak AJ (2002) IBD (Isolation By Distance): a program for analyses of isolation by distance. J Heredity 93:153–154
Borrell AK, Hammer GL, Douglas ACL (2000) Does maintaining green leaf area in sorghum improve yield under drought? I. Leaf growth and senescence. Crop Sci 40:11026–11103
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Human Genet 32:314–331
Brown SM, Hopkins MS, Mitchell SE, Senior ML, Wang TY, Duncan RR, Gonzalez-Candelas F, Kresovich S (1996) Multiple methods for the identification of polymorphic simple sequence repeats (SSRs) in sorghum [Sorghum bicolor (L.) Moench]. Theor Appl Genet 93:190–198
Casa AM, Mitchell SE, Hamblin MT, Sun H, Bowers JE, Paterson AH, Aquadro CF, Kresovich S (2005) Diversity and selection in sorghum: simultaneous analyses using simple sequence repeats. Theor Appl Genet 111:23–30
Central Statistical Agency (CSA) (2010) Agricultural sample survey, report on area and production of crops (private peasant holdings, Meher season), vol IV. Addis Ababa, Ethiopia
Chakraborty R, Jin L (1993) Determination of relatedness between individuals by DNA fingerprinting. Human Biol 65:875–895
Cui YX, Xu GX, Magill CW, Schertz KF, Hart GE (1995) RFLP-based assay of Sorghum bicolor (L.) Moench genetic diversity. Theor Appl Genet 90:787–796
de Wet JMJ (1978) Systematics and evolution of sorghum sect. Sorghum (Gramineae). Am J Bot 65:477–484
de Wet JMJ, Harlan JR (1971) The origin and domestication of Sorghum bicolor. Econ Bot 25:128–131
Dean RE, Dahlberg JA, Hopkins MS, Mitchell SE, Kresovich S (1999) Genetic redundancy and diversity among ‘orange’ accessions in the U.S. national sorghum collection as assessed with simple sequence repeat (SSR) markers. Crop Sci 39:1215–1222
Djé Y, Heuertz M, Lefebvre C, Vekemans X (2000) Assessment of genetic diversity within and among germplasm accessions in cultivated sorghum using microsatellite markers. Theor Appl Genet 100:918–925
Djé Y, Heuertz M, Ater M, Lefebyre C, Vekemans X (2004) In situ estimation of outcrossing rate in sorghum landraces using microsatellite markers. Euphytica 138:205–212
Doggett H (1988) Sorghum, 2nd edn. Longman, London
Doggett H, Majisu BN (1968) Disruptive selection in crop development. Heredity 23:1–23
Earl and von Holdt (2011) STRUCTURE HARVESTER v0.6.8. http://taylor0.biology.ucla.edu/struct_harvest). Accessed 2 Nov 2011
Ejeta G, Grenier C (2005) Sorghum and its weedy hybrids. In: Gressel J (ed) Crop ferality and volunteerism. Taylor and Francis, Florida, pp 123–135
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Ann Rev Ecol Syst 24:217–242
Ellstrand NC, Prentice HC, Hancock JF (1999) Gene flow and introgression from domesticated plants into their wild relatives. Ann Rev Ecol Evol Syst 30:539–563
Escalante A, Coello MG, Eguiarte LE, Piñero D (1994) Genetic structure and mating systems in wild and cultivated populations of Phaseolus coccineus and P. vulgaris (Fabaceae). Am J Bot 81:1096–1103
Evanno S, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinfo Online 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Ghebru B, Schmidt RJ, Bennetzen JL (2002) Genetic diversity of Eritrean sorghum landraces assessed with simple sequence repeat (SSR) markers. Theor Appl Genet 105:229–236
Goudet J (2002) Fstat, version 2.932. In: Institute of Ecology, Biology Building, UNIL, Switzerland, Lausanne
Harlan JR, de Wet JMJ (1972) A simplified classification of cultivated plants. Taxon 20:509–517
Heywooda VH, Iriondob JM (2003) Plant conservation: old problems, new perspectives. Biol Conserv 113:321–335
Hulbert SH (1971) The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577–586
Idury RM, Cardon LR (1997) A simple method for automated allele binning in microsatellite markers. Genome Res 11:1104–1109
Jarvis DI, Hodgkin T (1999) Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agroecosystems. Mol Ecol 8:5159–5173
Kalinowski S (2005) HP-RARE 10: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol 5:187–189
Ladizinsky G (1985) Founder effect in crop-plant evolution. Econ Bot 39:191–199
Langridge P, Chalmers K (2004) The principle: identification and application of molecular markers. In: Lörz H, Wenzel G (eds) Molecular marker systems in plant breeding and crop improvement. Biotechnology in Agriculture and Forestry, vol 55. Springer, Berlin, pp 3–22
Li M, Yuyama N, Luo L, Hirata M, Cai H (2009) In silico mapping of 1758 new SSR markers developed from public genomic sequences for sorghum. Mol Breed 24:41–47
Liang H, Gao ZS (2001) Phylogenetic analysis and transformation of sorghum [Sorghum bicolor (L.) Moench]. Recent Res Dev Plant Biol 1:17–33
Liu J (2005) PowerMarker V3.125. http://www.powermarker.net. Accessed 21 Aug 2009
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Maxted N (2003) Conserving the genetic resources of crop wild relatives in European Protected Areas. Biol Conserv 113:411–417
Morden CW, Doebley JF, Schertz KF (1989) Allozyme variation in old world races of Sorghum bicolor (Poaceae). Am J Bot 76:247–255
Morden CW, Doebley JF, Schertz KF (1990) Allozyme variation among the spontaneous species of Sorghum section Sorghum (Poaceae). Theor Appl Genet 80:296–304
Muraya MM, Mutegi E, Geiger HH, de Villiers SM, Sagnard F, Kanyenji BM, Kiambi D, Parzies HK (2011) Wild sorghum from different eco-geographic regions of Kenya display a mixed mating system. Theor Appl Genet 122:1631–1639. doi:10.1007/s00122-011-1560-5
Mutegi E, Sagnard F, Muraya M, Kanyenji B, Rono B, Mwongera C, Marangu C, Kamau J, Parzies H, de Villiers S, Semagn K, Traore’ PS, Labuschagne M (2010) Ecogeographical distribution of wild weedy and cultivated Sorghum bicolor (L.) Moench in Kenya: implications for conservation and crop-to-wild gene flow. Genet Resour Crop Evol 57:243–253
Mutegi E, Sagnard F, Semagn K, Deu M, Muraya M, Kanyenji B, de Villiers S, Kiambi D, Herselman L, Labuschagne M (2011) Genetic structure and relationships within and between cultivated and wild sorghum (Sorghum bicolor (L.) Moench) in Kenya as revealed by microsatellite markers. Theor Appl Genet 122:989–1004
Mutegi E, Sagnard F, Labuschagne M, Herselman L, Semagn K, Deu M, de Villiers S, Kanyenji BM, Mwongera CN, Traore PCS, Kiambi D (2012) Local scale patterns of gene flow and genetic diversity in a crop–wild–weedy complex of sorghum (Sorghum bicolor (L.) Moench) under traditional agricultural field conditions in Kenya. Conserv Genet. doi:10.1007/s10592-012-0353-y
Page RDM (2001) TreeView 1.6.6. http://taxonomy.zoology.gla.ac.uk/rod/rod.html
Pedersen JF, Toy JJ, Johnson B (1998) Natural outcrossing of sorghum and sudangrass in the central great plains. Crop Sci 38:937–939
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin
Prasanth V, Chandra S, Jayashree B, Hoisington D (2006) AlleloBin-a program for allele binning of microsatellite markers based on the algorithm of Idury and Cardon (1997). International Crops Research Institute for the Semi, Arid Tropics (ICRISAT), Patancheru, AP, India
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN:3-900051-07-0, URL http://www.R-project.org
Reed JD, Ramundo BA, Claflin LE, Tuinstra MR (2002) Analysis of resistance to ergot in sorghum and potential alternate hosts. Crop Sci 42:1135–1138
Rice EB (2004) Conservation and change: a comparison of in-situ and ex-situ conservation of Jala maize germplasm in Mexico. PhD Dissertation, Cornell University
Rousset F, Raymond M (1997) Statistical analyses of population genetic data: new tools, old concepts. Trends Ecol Evol 12:313–317
Sagnard F, Deu M, Dembe’le’ D, Leblois R, Toure’L, Diakite’ M, Calatayud C, Vaksmann M, Bouchet S, Malle’ Y, Togola S, Traore PCS (2011) Genetic diversity, structure, gene flow and evolutionary relationships within the Sorghum bicolor wild–weedy–crop complex in a western African region. Theor Appl Genet 123:1231–1246
Schmidt MR, Bothma GC (2006) Risk sssessment for transgenic sorghum in Africa: crop-to-crop gene flow in Sorghum bicolor (L.) Moench. Crop Sci 46:790–798
Seboka B, van Hintum T (2006) The dynamics of on-farm management of sorghum in Ethiopia: implication for the conservation and improvement of plant genetic resources. Genet Resour Crop Evol 53:1385–1403
Singh R, Axtell JD (1973) High lysine mutant gene (hl) that improves protein quality and biological value of grain sorghum. Crop Sci 13:535
Small E (1984) Hybridization in the domesticated-weed-wild complex. In: Grant WF (ed) Plant biosystematics. Academic Press, Toronto, pp 195–210
Snow AA, Andow DA, Gepts P, Hallerman EM, Power A, Tiedje JM, Wolfenbarger LL (2005) Genetically engineered organisms and the environment: current status and recommendations. Ecol Appl 15:377–404
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. doi:10.1093/bioinformatics/btn478
Tanksley SD, McCouch R (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Taramino G, Tarchini R, Ferrario S, Lee M, Pe ME (1997) Characterization and mapping of simple sequence repeats (SSRs) in Sorghum bicolor. Theor Appl Genet 95:66–72
Templeton AR (2006) Population genetics and microevolutionary theory. Wiley, New Jersey
Teshome A, Baum BR, Fahrig L, Torrance JK, Arnason TJ, Lambert JD (1997) Sorghum [Sorghum bicolor (L.) Moench] landrace variation and classification in North Shewa and SouthWelo, Ethiopia. Euphytica 97:255–263
Tesso T, Kapran I, Grenier C, Snow AA, Sweeney P, Pedersen J, Marx D, Bothma G, Ejeta G (2008) The potential for crop-to-wild gene flow in sorghum in Ethiopia and Niger: a geographic survey. Crop Sci 48:1425–1431
Weir S, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Wu YQ, Huang Y, Tauer CG, Porter DR (2006) Genetic diversity of sorghum accessions resistant to greenbugs as assessed with AFLP markers. Genome 49:143–149
Volis S, Blecher M, Sapir Y (2009) Complex ex situ—in situ approach for conservation of endangered plant species and its application to Iris atrofusca of the Northern Negev. In: Krupp F, Musselman LJ, Kotb MMA, Weidig I (Eds) Environment, biodiversity and conservation in the middle east, vol 3. BioRisk, pp 137–160
Zhao Z (2007) The Africa biofortified sorghum project –applying biotechnology to develop nutritionally improved sorghum for Africa. In: Xu Z et al (eds) Biotechnology and sustainable agriculture 2006 and beyond. Springer, Berlin, pp 273–277
Zhao ZY, Cai T, Tagliani L, Miller M, Wang N, Pang H, Rudert M, Schroeder S, Hondred D, Seltzer J, Pierce D (2000) Agrobacterium mediated sorghum transformation. Plant Mol Biol 44:789–798
Zhao ZY, Glassman K, Sewalt V, Wang N, Miller M, Chang S, Thompson T, Catron S, Wu E, Bidney D, Kedebe Y, Jung R (2003) Nutritionally improved transgenic sorghum. In: Vasil IK (ed) Plant biotechnology 2002 and beyond. Kluwer Academic Publishers, Florida, pp 413–416
Zhu H, Muthukrishnan S, Krishnaveni S, Wilde G, Jeoung JM, Liang GH (1998) Biolistic transformation of sorghum using a rice chitinase gene. J Genet Breed 52:243–252
Acknowledgments
This study was supported by a grant from the Biotechnology-Biodiversity Interface (BBI) program of the United States Agency for International Development (USAID) to A. A. Snow and serves as part of the dissertation research for the partial fulfillment of the requirements for a PhD degree to the first author. We also thank G. Ejeta, C. Grenier, J. Pederson, T. Tesso, and Y. Teklu for helpful advice and assistance with this research. We extend our sincere thanks to ICRISAT for providing the seeds of wild sorghum accessions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Adugna, A., Snow, A.A., Sweeney, P.M. et al. Population genetic structure of in situ wild Sorghum bicolor in its Ethiopian center of origin based on SSR markers. Genet Resour Crop Evol 60, 1313–1328 (2013). https://doi.org/10.1007/s10722-012-9921-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-012-9921-8